Conservation and divergence of protein pathways in the vertebrate heart
- PMID: 31490923
- PMCID: PMC6750614
- DOI: 10.1371/journal.pbio.3000437
Conservation and divergence of protein pathways in the vertebrate heart
Abstract
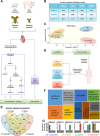
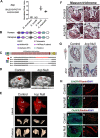
Heart disease is the leading cause of death in the western world. Attaining a mechanistic understanding of human heart development and homeostasis and the molecular basis of associated disease states relies on the use of animal models. Here, we present the cardiac proteomes of 4 model vertebrates with dual circulatory systems: the pig (Sus scrofa), the mouse (Mus musculus), and 2 frogs (Xenopus laevis and Xenopus tropicalis). Determination of which proteins and protein pathways are conserved and which have diverged within these species will aid in our ability to choose the appropriate models for determining protein function and to model human disease. We uncover mammalian- and amphibian-specific, as well as species-specific, enriched proteins and protein pathways. Among these, we find and validate an enrichment in cell-cycle-associated proteins within Xenopus laevis. To further investigate functional units within cardiac proteomes, we develop a computational approach to profile the abundance of protein complexes across species. Finally, we demonstrate the utility of these data sets for predicting appropriate model systems for studying given cardiac conditions by testing the role of Kielin/chordin-like protein (Kcp), a protein found as enriched in frog hearts compared to mammals. We establish that germ-line mutations in Kcp in Xenopus lead to valve defects and, ultimately, cardiac failure and death. Thus, integrating these findings with data on proteins responsible for cardiac disease should lead to the development of refined, species-specific models for protein function and disease states.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Heron M, Hoyert DL, Murphy SL, Xu J, Kochanek KD, Tejada-Vera B. Deaths: final data for 2006. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2009;57(14):1–134. Epub 2009/10/01. . - PubMed
-
- Oikonomopoulos A, Kitani T, Wu JC. Pluripotent Stem Cell-Derived Cardiomyocytes as a Platform for Cell Therapy Applications: Progress and Hurdles for Clinical Translation. Molecular therapy: the journal of the American Society of Gene Therapy. 2018;26(7):1624–34. Epub 2018/04/28. 10.1016/j.ymthe.2018.02.026 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

