Molecular epidemiology of carbapenem-resistance plasmids using publicly available sequences
- PMID: 31491336
- PMCID: PMC9221968
- DOI: 10.1139/gen-2019-0100
Molecular epidemiology of carbapenem-resistance plasmids using publicly available sequences
Abstract
Carbapenem-resistant bacteria have quickly become a worldwide concern in nosocomial infections. Of the seven known carbapenemases, four have been shown to be particularly problematic: KPC, NDM, IMP, and VIM. To date, many local and species- or carbapenemase-specific epidemiological studies have been performed, which often focus on the organism itself. This report attempts to perform an inclusive (encompass both species and carbapenemase) epidemiologic study using publicly available plasmid sequences from NCBI. In this report, the gene content of these various plasmids has been characterized, replicon types of the plasmids identified, and the global spread and species promiscuity of the plasmids analyzed. Additionally, support to several groups targeting plasmid maintenance and transfer mechanisms to slow the spread of resistance plasmids is given.
Les bactéries résistantes aux carbapénèmes sont rapidement devenues un problème mondial en matière d’infections nosocomiales. Des sept carbapénémases connues, quatre se sont montrées particulièrement problématiques : KPC, NDM, IMP et VIM. À ce jour, plusieurs études épidémiologiques à portée locale ou spécifiques d’une espèce ou d’une carbapénémase ont été réalisées et portaient généralement sur l’organisme lui-même. Ce travail cherchait à réaliser une étude épidémiologique englobante (incluant à la fois les espèces et les carbapénémases) à partir des données publiques sur les séquences plasmidiques disponibles dans NCBI. Dans ce travail, les auteurs rapportent le contenu en gènes des différents plasmides qui ont été caractérisés, les types de réplicons chez les plasmides identifiés ainsi que la propagation mondiale et la promiscuité des espèces pour les plasmides analysés. De plus, un appui est fourni aux nombreux groupes qui travaillent sur le maintien des plasmides et leurs mécanismes de transfert afin de ralentir la propagation des plasmides conférant la résistance. [Traduit par la Rédaction]
Keywords: Enterobacteriaceae; antibiotic resistance; carbapenemase; carbapénémase; distribution mondiale; global distribution; plasmid; plasmide; résistance aux antimicrobiens.
Conflict of interest statement
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
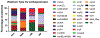
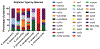

Similar articles
-
Spreading Patterns of NDM-Producing Enterobacteriaceae in Clinical and Environmental Settings in Yangon, Myanmar.Antimicrob Agents Chemother. 2019 Feb 26;63(3):e01924-18. doi: 10.1128/AAC.01924-18. Print 2019 Mar. Antimicrob Agents Chemother. 2019. PMID: 30530602 Free PMC article.
-
Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789).BMC Microbiol. 2015 Nov 9;15(1):260. doi: 10.1186/s12866-015-0597-z. BMC Microbiol. 2015. PMID: 26552763 Free PMC article.
-
Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review.Ann N Y Acad Sci. 2019 Dec;1457(1):61-91. doi: 10.1111/nyas.14223. Epub 2019 Aug 30. Ann N Y Acad Sci. 2019. PMID: 31469443 Review.
-
Enhanced Carbapenem Resistance through Multimerization of Plasmids Carrying Carbapenemase Genes.mBio. 2021 Jun 29;12(3):e0018621. doi: 10.1128/mBio.00186-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154401 Free PMC article.
-
The rapid spread of carbapenem-resistant Enterobacteriaceae.Drug Resist Updat. 2016 Nov;29:30-46. doi: 10.1016/j.drup.2016.09.002. Epub 2016 Sep 19. Drug Resist Updat. 2016. PMID: 27912842 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

