Mendelian Gene Discovery: Fast and Furious with No End in Sight
- PMID: 31491408
- PMCID: PMC6731362
- DOI: 10.1016/j.ajhg.2019.07.011
Mendelian Gene Discovery: Fast and Furious with No End in Sight
Abstract
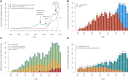
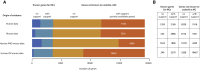
Gene discovery for Mendelian conditions (MCs) offers a direct path to understanding genome function. Approaches based on next-generation sequencing applied at scale have dramatically accelerated gene discovery and transformed genetic medicine. Finding the genetic basis of ∼6,000-13,000 MCs yet to be delineated will require both technical and computational innovation, but will rely to a larger extent on meaningful data sharing.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Antonarakis S.E., Beckmann J.S. Mendelian disorders deserve more attention. Nat. Rev. Genet. 2006;7:277–282. - PubMed
-
- Starita L.M., Islam M.M., Banerjee T., Adamovich A.I., Gullingsrud J., Fields S., Shendure J., Parvin J.D. A multiplex homology-directed DNA repair assay reveals the impact of more than 1,000 BRCA1 missense substitution variants on protein function. Am. J. Hum. Genet. 2018;103:498–508. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

