Olfactory receptor genes make the case for inter-chromosomal interactions
- PMID: 31491591
- PMCID: PMC6759391
- DOI: 10.1016/j.gde.2019.07.004
Olfactory receptor genes make the case for inter-chromosomal interactions
Abstract
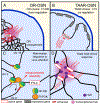
The partitioning of the interphase nucleus into chromosome territories generally precludes DNA from making specific and reproducible inter-chromosomal contacts. However, with the development of powerful genomic and imaging tools for the analysis of the 3D genome, and with their application on an increasing number of cell types, it becomes apparent that regulated, specific, and functionally important inter-chromosomal contacts exist. Widespread and stereotypic inter-chromosomal interactions are at the center of chemosensation, where they regulate the singular and stochastic expression of olfactory receptor genes. In olfactory sensory neurons (OSNs) coalescence of multiple intergenic enhancers to a multi-chromosomal hub orchestrates the expression of a single OR allele, whereas convergence of the remaining OR genes from 18 chromosomes into a few heterochromatic compartments mediates their effective transcriptional silencing. In this review we describe the role of interchromosomal interactions in OR gene choice, and we describe other biological systems where such genomic interactions may contribute to regulatory robustness and transcriptional diversification.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures


References
-
- Cremer T, Cremer C et al. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments.Hum Genet 60(1),46–56 (1982). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

