Finding maximally disconnected subnetworks with shortest path tractography
- PMID: 31491834
- PMCID: PMC6627647
- DOI: 10.1016/j.nicl.2019.101903
Finding maximally disconnected subnetworks with shortest path tractography
Abstract
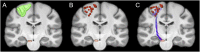
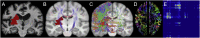
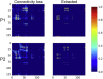
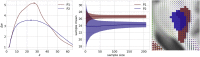
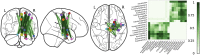
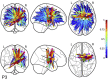
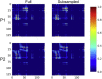
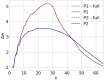
Connectome-based lesion symptom mapping (CLSM) can be used to relate disruptions of brain network connectivity with clinical measures. We present a novel method that extends current CLSM approaches by introducing a fast reliable and accurate way for computing disconnectomes, i.e. identifying damaged or lesioned connections. We introduce a new algorithm that finds the maximally disconnected subgraph containing regions and region pairs with the greatest shared connectivity loss. After normalizing a stroke patient's segmented MRI lesion into template space, probability weighted structural connectivity matrices are constructed from shortest paths found in white matter voxel graphs of 210 subjects from the Human Connectome Project. Percent connectivity loss matrices are constructed by measuring the proportion of shortest-path probability weighted connections that are lost because of an intersection with the patient's lesion. Maximally disconnected subgraphs of the overall connectivity loss matrix are then derived using a computationally fast greedy algorithm that closely approximates the exact solution. We illustrate the approach in eleven stroke patients with hemiparesis by identifying expected disconnections of the corticospinal tract (CST) with cortical sensorimotor regions. Major disconnections are found in the thalamus, basal ganglia, and inferior parietal cortex. Moreover, the size of the maximally disconnected subgraph quantifies the extent of cortical disconnection and strongly correlates with multiple clinical measures. The methods provide a fast, reliable approach for both visualizing and quantifying the disconnected portion of a patient's structural connectome based on their routine clinical MRI, without reliance on concomitant diffusion weighted imaging. The method can be extended to large databases of stroke patients, multiple sclerosis or other diseases causing focal white matter injuries helping to better characterize clinically relevant white matter lesions and to identify biomarkers for the recovery potential of individual patients.
Keywords: Brain injury; Brain networks; Connectomes; Diffusion MRI; Disconnection; Disconnectome; Graphs; Lesion symptom mapping; Spatial normalization; Tractography.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Binkofski F., Seitz R.J., Hackländer T., Pawelec D., Mau J., Freund H.J. Recovery of motor functions following hemiparetic stroke: a clinical and magnetic resonance-morphometric study. Cerebrovasc. Dis. 2001;11:273–281. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

