Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells
- PMID: 31491855
- PMCID: PMC6770266
- DOI: 10.3390/ijms20184345
Functional Dissection of pri-miR-290~295 in Dgcr8 Knockout Mouse Embryonic Stem Cells
Abstract
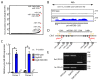
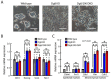
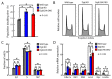
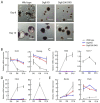
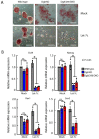
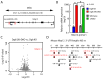
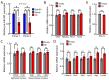
The DiGeorge syndrome critical region gene 8 (Dgcr8) knockout strategy has been widely used to study the function of canonical microRNAs (miRNAs) in vitro and in vivo. However, primary miRNA (pri-miRNA) transcripts are accumulated in Dgcr8 knockout cells due to interrupted processing. Whether abnormally accumulated pri-miRNAs have any function is unknown. Here, using clustered regularly interspaced short palindromic repeats system/CRISPR-associated protein 9 (CRISPR/Cas9), we successfully knocked out the primary microRNA-290~295 (pri-miR-290~295) cluster, the most highly expressed miRNA cluster in mouse embryonic stem cells (ESCs), in Dgcr8 knockout background. We found that the major defects associated with Dgcr8 knockout in mouse ESCs, including higher expression of epithelial-to-mesenchymal transition (EMT) markers, slower proliferation, G1 accumulation, and defects in silencing self-renewal, were not affected by the deletion of pri-miR-290~290 cluster. Interestingly, the transcription of neighboring gene nucleotide-binding oligomerization domain, leucine rich repeat and pyrin domain containing 12(Nlrp12) was upregulated upon the deletion of the pri-miR-290~295 cluster. Together, our results suggested that the major defects in Dgcr8 knockout ESCs were not due to the accumulation of pri-miR-290~295, and the deletion of miRNA genes could affect the transcription of neighboring DNA elements.
Keywords: DiGeorge syndrome critical region gene 8; Nlrp12; cell cycle; differentiation; embryonic stem cells; miR-290~295 cluster; pluripotency; primary microRNA; self-renewal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

