A Parallel Phenotypic Versus Target-Based Screening Strategy for RNA-Dependent RNA Polymerase Inhibitors of the Influenza A Virus
- PMID: 31491939
- PMCID: PMC6783926
- DOI: 10.3390/v11090826
A Parallel Phenotypic Versus Target-Based Screening Strategy for RNA-Dependent RNA Polymerase Inhibitors of the Influenza A Virus
Abstract
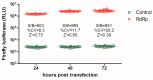
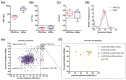
Influenza A virus infections cause significant morbidity and mortality, and novel antivirals are urgently needed. Influenza RNA-dependent RNA polymerase (RdRp) activity has been acknowledged as a promising target for novel antivirals. In this study, a phenotypic versus target-based screening strategy was established to identify the influenza A virus inhibitors targeting the virus RNA transcription/replication steps by sequentially using an RdRp-targeted screen and a replication-competent reporter virus-based approach using the same compounds. To demonstrate the utility of this approach, a pilot screen of a library of 891 compounds derived from natural products was carried out. Quality control analysis indicates that the primary screen was robust for identification of influenza A virus inhibitors targeting RdRp activity. Finally, two hit candidates were identified, and one was validated as a putative RdRp inhibitor. This strategy can greatly reduce the number of false positives and improve the accuracy and efficacy of primary screening, thereby providing a powerful tool for antiviral discovery.
Keywords: Influenza A virus; RNA dependent RNA polymerase inhibitor; phenotypic screen; target-based screen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A small molecule multi-kinase inhibitor reduces influenza A virus replication by restricting viral RNA synthesis.Antiviral Res. 2013 Oct;100(1):29-37. doi: 10.1016/j.antiviral.2013.07.010. Epub 2013 Jul 24. Antiviral Res. 2013. PMID: 23891991
-
Influenza A virus polymerase: an attractive target for next-generation anti-influenza therapeutics.Drug Discov Today. 2018 Mar;23(3):503-518. doi: 10.1016/j.drudis.2018.01.028. Epub 2018 Jan 12. Drug Discov Today. 2018. PMID: 29339107 Review.
-
Neuraminidase activity provides a practical read-out for a high throughput influenza antiviral screening assay.Virol J. 2008 Sep 26;5:109. doi: 10.1186/1743-422X-5-109. Virol J. 2008. PMID: 18822145 Free PMC article.
-
Zinc Finger-Containing Cellular Transcription Corepressor ZBTB25 Promotes Influenza Virus RNA Transcription and Is a Target for Zinc Ejector Drugs.J Virol. 2017 Sep 27;91(20):e00842-17. doi: 10.1128/JVI.00842-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28768860 Free PMC article.
-
Focusing on the Influenza Virus Polymerase Complex: Recent Progress in Drug Discovery and Assay Development.Curr Med Chem. 2019;26(13):2243-2263. doi: 10.2174/0929867325666180706112940. Curr Med Chem. 2019. PMID: 29984646 Free PMC article. Review.
Cited by
-
Establishment and application of a high-throughput screening model for cell adhesion inhibitors.Front Pharmacol. 2023 Feb 23;14:1140163. doi: 10.3389/fphar.2023.1140163. eCollection 2023. Front Pharmacol. 2023. PMID: 36909195 Free PMC article.
-
Screening for Anti-Influenza Actives of Prefractionated Traditional Chinese Medicines.Evid Based Complement Alternat Med. 2020 Oct 14;2020:4979850. doi: 10.1155/2020/4979850. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 33123207 Free PMC article.
-
Semisynthetic Cardenolides Acting as Antiviral Inhibitors of Influenza A Virus Replication by Preventing Polymerase Complex Formation.Molecules. 2020 Oct 21;25(20):4853. doi: 10.3390/molecules25204853. Molecules. 2020. PMID: 33096707 Free PMC article.
-
Allopregnanolone targets nucleoprotein as a novel influenza virus inhibitor.Virol Sin. 2023 Dec;38(6):931-939. doi: 10.1016/j.virs.2023.09.003. Epub 2023 Sep 22. Virol Sin. 2023. PMID: 37741571 Free PMC article.
-
Identification of Chebulinic Acid and Chebulagic Acid as Novel Influenza Viral Neuraminidase Inhibitors.Front Microbiol. 2020 Feb 28;11:182. doi: 10.3389/fmicb.2020.00182. eCollection 2020. Front Microbiol. 2020. PMID: 32256457 Free PMC article.
References
-
- WHO . 2018 Influenza (Seasonal) Fact Sheet. WHO; Geneva, Switzerland: 2018.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

