Computational Study of Natural Compounds for the Clearance of Amyloid-Βeta: A Potential Therapeutic Management Strategy for Alzheimer's Disease
- PMID: 31491967
- PMCID: PMC6767296
- DOI: 10.3390/molecules24183233
Computational Study of Natural Compounds for the Clearance of Amyloid-Βeta: A Potential Therapeutic Management Strategy for Alzheimer's Disease
Abstract
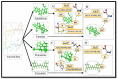
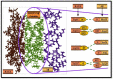
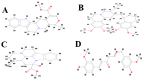
Alzheimer's disease (AD) is a widespread dynamic neurodegenerative malady. Its etiology is still not clear. One of the foremost pathological features is the extracellular deposits of Amyloid-beta (Aβ) peptides in senile plaques. The interaction of Aβ and the receptor for advanced glycation end products at the blood-brain barrier is also observed in AD, which not only causes the neurovascular anxiety and articulation of proinflammatory cytokines, but also directs reduction of cerebral bloodstream by upgrading the emission of endothelin-1 to induce vasoconstriction. In this process, RAGE is deemed responsible for the influx of Aβ into the brain through BBB. In the current study, we predicted the interaction potential of the natural compounds vincamine, ajmalicine and emetine with the Aβ peptide concerned in the treatment of AD against the standard control, curcumin, to validate the Aβ peptide-compounds results. Protein-protein interaction studies have also been carried out to see their potential to inhibit the binding process of Aβ and RAGE. Moreover, the current study verifies that ligands are more capable inhibitors of a selected target compared to positive control with reference to ΔG values. The inhibition of Aβ and its interaction with RAGE may be valuable in proposing the next round of lead compounds for effective Alzheimer's disease treatment.
Keywords: Alzheimer’s disease; Z-dock; binding energy; docking; natural compounds.
Conflict of interest statement
The authors confirm that this article content has no conflict of interest.
Figures


Similar articles
-
The ongoing search for small molecules to study metal-associated amyloid-β species in Alzheimer's disease.Acc Chem Res. 2014 Aug 19;47(8):2475-82. doi: 10.1021/ar500152x. Epub 2014 Jul 31. Acc Chem Res. 2014. PMID: 25080056
-
Screening and Elucidation of Selected Natural Compounds for Anti- Alzheimer's Potential Targeting BACE-1 Enzyme: A Case Computational Study.Curr Comput Aided Drug Des. 2017 Nov 10;13(4):311-318. doi: 10.2174/1573409913666170414123825. Curr Comput Aided Drug Des. 2017. PMID: 28413992
-
Amyloid binding ligands as Alzheimer's disease therapies.Neurobiol Aging. 2002 Nov-Dec;23(6):1039-42. doi: 10.1016/s0197-4580(02)00121-5. Neurobiol Aging. 2002. PMID: 12470800 Review.
-
Computational Insight into the Effect of Natural Compounds on the Destabilization of Preformed Amyloid-β(1⁻40) Fibrils.Molecules. 2018 May 31;23(6):1320. doi: 10.3390/molecules23061320. Molecules. 2018. PMID: 29857500 Free PMC article.
-
Alzheimer's disease.Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review.
Cited by
-
The pharmacokinetics profiles, pharmacological properties, and toxicological risks of dehydroevodiamine: A review.Front Pharmacol. 2022 Nov 18;13:1040154. doi: 10.3389/fphar.2022.1040154. eCollection 2022. Front Pharmacol. 2022. PMID: 36467053 Free PMC article. Review.
-
Protective effects of vincamine against ethanol-induced gastric ulcer by attenuation of IL-6, IL-1β, and TNF-α mRNA expression levels in the gastric mucosa of BALB/c mice.J Mol Histol. 2025 Mar 5;56(2):100. doi: 10.1007/s10735-025-10374-x. J Mol Histol. 2025. PMID: 40038147
-
Study of Caspase 8 Inhibition for the Management of Alzheimer's Disease: A Molecular Docking and Dynamics Simulation.Molecules. 2020 Apr 29;25(9):2071. doi: 10.3390/molecules25092071. Molecules. 2020. PMID: 32365525 Free PMC article.
-
Therapeutic potential of targeting the receptor for advanced glycation end products (RAGE) by small molecule inhibitors.Drug Dev Res. 2022 Sep;83(6):1257-1269. doi: 10.1002/ddr.21971. Epub 2022 Jul 4. Drug Dev Res. 2022. PMID: 35781678 Free PMC article. Review.
-
Identification of Persuasive Antiviral Natural Compounds for COVID-19 by Targeting Endoribonuclease NSP15: A Structural-Bioinformatics Approach.Molecules. 2020 Dec 1;25(23):5657. doi: 10.3390/molecules25235657. Molecules. 2020. PMID: 33271751 Free PMC article.
References
-
- Alzheimer’s Association Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–384. - PubMed
-
- Kayed R., Sokolov Y., Edmonds B., McIntire T.M., Milton S.C., Hall J.E., Glabe C.G. Permeabilization of Lipid Bilayers Is a Common Conformation-dependent Activity of Soluble Amyloid Oligomers in Protein Misfolding Diseases. J. Boil. Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

