Multi-omics signature of brain amyloid deposition in asymptomatic individuals at-risk for Alzheimer's disease: The INSIGHT-preAD study
- PMID: 31492558
- PMCID: PMC6796577
- DOI: 10.1016/j.ebiom.2019.08.051
Multi-omics signature of brain amyloid deposition in asymptomatic individuals at-risk for Alzheimer's disease: The INSIGHT-preAD study
Abstract
Background: One of the biggest challenge in Alzheimer's disease (AD) is to identify pathways and markers of disease prediction easily accessible, for prevention and treatment. Here we analysed blood samples from the INveStIGation of AlzHeimer's predicTors (INSIGHT-preAD) cohort of elderly asymptomatic individuals with and without brain amyloid load.
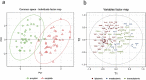
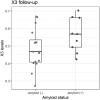
Methods: We performed blood RNAseq, and plasma metabolomics and lipidomics using liquid chromatography-mass spectrometry on 48 individuals amyloid positive and 48 amyloid negative (SUVr cut-off of 0·7918). The three data sets were analysed separately using differential gene expression based on negative binomial distribution, non-parametric (Wilcoxon) and parametric (correlation-adjusted Student't) tests. Data integration was conducted using sparse partial least squares-discriminant and principal component analyses. Bootstrap-selected top-ten features from the three data sets were tested for their discriminant power using Receiver Operating Characteristic curve. Longitudinal metabolomic analysis was carried out on a subset of 22 subjects.
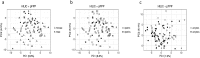
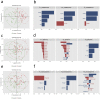
Findings: Univariate analyses identified three medium chain fatty acids, 4-nitrophenol and a set of 64 transcripts enriched for inflammation and fatty acid metabolism differentially quantified in amyloid positive and negative subjects. Importantly, the amounts of the three medium chain fatty acids were correlated over time in a subset of 22 subjects (p < 0·05). Multi-omics integrative analyses showed that metabolites efficiently discriminated between subjects according to their amyloid status while lipids did not and transcripts showed trends. Finally, the ten top metabolites and transcripts represented the most discriminant omics features with 99·4% chance prediction for amyloid positivity.
Interpretation: This study suggests a potential blood omics signature for prediction of amyloid positivity in asymptomatic at-risk subjects, allowing for a less invasive, more accessible, and less expensive risk assessment of AD as compared to PET studies or lumbar puncture. FUND: Institut Hospitalo-Universitaire and Institut du Cerveau et de la Moelle Epiniere (IHU-A-ICM), French Ministry of Research, Fondation Alzheimer, Pfizer, and Avid.
Keywords: Alzheimer; Amyloid PET; Asymptomatic; Biomarkers; Multi-omics; Prediction.
Copyright © 2019. Published by Elsevier B.V.
Conflict of interest statement
BD has received consultancy fees from Biogen, Boehringer Ingelheim and Eli Lilly, and grants for his institution from Merck, Pfizer, and Roche. SE has received grants from Eli Lilly and consul tant fees from Astellas Pharma. M-CP has received grants from Fondation Vaincre Alzheimer, Laboratoires Servier, Pfizer, and Roche. M-OH has received consultant fees from Eli Lilly and speaker fees from Piramal. The other authors declare no competing interests.
Figures





References
-
- Golde T.E., DeKosky S.T., Galasko D. Alzheimer's disease: the right drug, the right time. Science. 2018;362(6420):1250–1251. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

