Potential Interactions between Clade SUP05 Sulfur-Oxidizing Bacteria and Phages in Hydrothermal Vent Sponges
- PMID: 31492669
- PMCID: PMC6821954
- DOI: 10.1128/AEM.00992-19
Potential Interactions between Clade SUP05 Sulfur-Oxidizing Bacteria and Phages in Hydrothermal Vent Sponges
Abstract
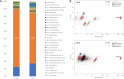
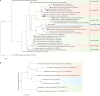
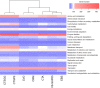
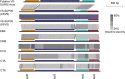
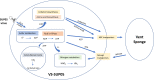
In deep-sea hydrothermal vent environments, sulfur-oxidizing bacteria belonging to the clade SUP05 are crucial symbionts of invertebrate animals. Marine viruses, as the most abundant biological entities in the ocean, play essential roles in regulating the sulfur metabolism of the SUP05 bacteria. To date, vent sponge-associated SUP05 and their phages have not been well documented. The current study analyzed microbiomes of Haplosclerida sponges from hydrothermal vents in the Okinawa Trough and recovered the dominant SUP05 genome, designated VS-SUP05. Phylogenetic analysis showed that VS-SUP05 was closely related to endosymbiotic SUP05 strains from mussels living in deep-sea hydrothermal vent fields. Homology and metabolic pathway comparisons against free-living and symbiotic SUP05 strains revealed that the VS-SUP05 genome shared many features with the deep-sea mussel symbionts. Supporting a potentially symbiotic lifestyle, the VS-SUP05 genome contained genes involved in the synthesis of essential amino acids and cofactors that are desired by the host. Analysis of sponge-associated viral sequences revealed putative VS-SUP05 phages, all of which were double-stranded viruses belonging to the families Myoviridae, Siphoviridae, Podoviridae, and Microviridae Among the phage sequences, one contig contained metabolic genes (iscR, iscS, and iscU) involved in iron-sulfur cluster formation. Interestingly, genome sequence comparison revealed horizontal transfer of the iscS gene among phages, VS-SUP05, and other symbiotic SUP05 strains, indicating an interaction between marine phages and SUP05 symbionts. Overall, our findings confirm the presence of SUP05 bacteria and their phages in sponges from deep-sea vents and imply a beneficial interaction that allows adaptation of the host sponge to the hydrothermal vent environment.IMPORTANCE Chemosynthetic SUP05 bacteria dominate the microbial communities of deep-sea hydrothermal vents around the world, SUP05 bacteria utilize reduced chemical compounds in vent fluids and commonly form symbioses with invertebrate organisms. This symbiotic relationship could be key to adapting to such unique and extreme environments. Viruses are the most abundant biological entities on the planet and have been identified in hydrothermal vent environments. However, their interactions with the symbiotic microbes of the SUP05 clade, along with their role in the symbiotic system, remain unclear. Here, using metagenomic sequence-based analyses, we determined that bacteriophages may support metabolism in SUP05 bacteria and play a role in the sponge-associated symbiosis system in hydrothermal vent environments.
Keywords: SUP05 bacteria; deep sea; hydrothermal vent; marine phages; sponge.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Newton ILG, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC, Fontanez KM, Lau E, Stewart FJ, Richardson PM, Barry KW, Saunders E, Detter JC, Wu D, Eisen JA, Cavanaugh CM. 2007. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315:998–1000. doi: 10.1126/science.1138438. - DOI - PubMed
-
- Ikuta T, Takaki Y, Nagai Y, Shimamura S, Tsuda M, Kawagucci S, Aoki Y, Inoue K, Teruya M, Satou K, Teruya K, Shimoji M, Tamotsu H, Hirano T, Maruyama T, Yoshida T. 2016. Heterogeneous composition of key metabolic gene clusters in a vent mussel symbiont population. ISME J 10:990–1001. doi: 10.1038/ismej.2015.176. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

