Hybrid Two-Component Sensors for Identification of Bacterial Chemoreceptor Function
- PMID: 31492670
- PMCID: PMC6821969
- DOI: 10.1128/AEM.01626-19
Hybrid Two-Component Sensors for Identification of Bacterial Chemoreceptor Function
Abstract
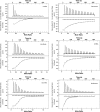
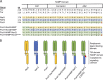
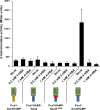
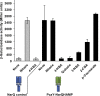
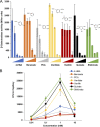
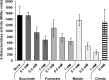
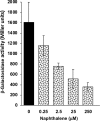
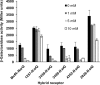
Soil bacteria adapt to diverse and rapidly changing environmental conditions by sensing and responding to environmental cues using a variety of sensory systems. Two-component systems are a widespread type of signal transduction system present in all three domains of life and typically are comprised of a sensor kinase and a response regulator. Many two-component systems function by regulating gene expression in response to environmental stimuli. The bacterial chemotaxis system is a modified two-component system with additional protein components and a response that, rather than regulating gene expression, involves behavioral adaptation and results in net movement toward or away from a chemical stimulus. Soil bacteria generally have 20 to 40 or more chemoreceptors encoded in their genomes. To simplify the identification of chemoeffectors (ligands) sensed by bacterial chemoreceptors, we constructed hybrid sensor proteins by fusing the sensor domains of Pseudomonas putida chemoreceptors to the signaling domains of the Escherichia coli NarX/NarQ nitrate sensors. Responses to potential attractants were monitored by β-galactosidase assays using an E. coli reporter strain in which the nitrate-responsive narG promoter was fused to lacZ Hybrid receptors constructed from PcaY, McfR, and NahY, which are chemoreceptors for aromatic acids, tricarboxylic acid cycle intermediates, and naphthalene, respectively, were sensitive and specific for detecting known attractants, and the β-galactosidase activities measured in E. coli correlated well with results of chemotaxis assays in the native P. putida strain. In addition, a screen of the hybrid receptors successfully identified new ligands for chemoreceptor proteins and resulted in the identification of six receptors that detect propionate.IMPORTANCE Relatively few of the thousands of chemoreceptors encoded in bacterial genomes have been functionally characterized. More importantly, although methyl-accepting chemotaxis proteins, the major type of chemoreceptors present in bacteria, are easily identified bioinformatically, it is not currently possible to predict what chemicals will bind to a particular chemoreceptor. Chemotaxis is known to play roles in biodegradation as well as in host-pathogen and host-symbiont interactions, but many studies are currently limited by the inability to identify relevant chemoreceptor ligands. The use of hybrid receptors and this simple E. coli reporter system allowed rapid and sensitive screening for potential chemoeffectors. The fusion site chosen for this study resulted in a high percentage of functional hybrids, indicating that it could be used to broadly test chemoreceptor responses from phylogenetically diverse samples. Considering the wide range of chemical attractants detected by soil bacteria, hybrid receptors may also be useful as sensitive biosensors.
Keywords: Pseudomonas putida; biosensor; methyl-accepting chemotaxis protein; receptor; two-component system.
Copyright © 2019 American Society for Microbiology.
Figures
Similar articles
-
Chemotaxis of Pseudomonas putida F1 to Alcohols Is Mediated by the Carboxylic Acid Receptor McfP.Appl Environ Microbiol. 2019 Oct 30;85(22):e01625-19. doi: 10.1128/AEM.01625-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31471307 Free PMC article.
-
Heterologous Expression of Pseudomonas putida Methyl-Accepting Chemotaxis Proteins Yields Escherichia coli Cells Chemotactic to Aromatic Compounds.Appl Environ Microbiol. 2018 Aug 31;84(18):e01362-18. doi: 10.1128/AEM.01362-18. Print 2018 Sep 15. Appl Environ Microbiol. 2018. PMID: 30006400 Free PMC article.
-
The expression of many chemoreceptor genes depends on the cognate chemoeffector as well as on the growth medium and phase.Curr Genet. 2017 Jun;63(3):457-470. doi: 10.1007/s00294-016-0646-7. Epub 2016 Sep 8. Curr Genet. 2017. PMID: 27632030
-
Bacterial chemoreceptors and chemoeffectors.Cell Mol Life Sci. 2015 Feb;72(4):691-708. doi: 10.1007/s00018-014-1770-5. Epub 2014 Nov 6. Cell Mol Life Sci. 2015. PMID: 25374297 Free PMC article. Review.
-
Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update.Trends Microbiol. 2015 May;23(5):257-66. doi: 10.1016/j.tim.2015.03.003. Epub 2015 Mar 30. Trends Microbiol. 2015. PMID: 25834953 Free PMC article. Review.
Cited by
-
Multiple functions of flagellar motility and chemotaxis in bacterial physiology.FEMS Microbiol Rev. 2021 Nov 23;45(6):fuab038. doi: 10.1093/femsre/fuab038. FEMS Microbiol Rev. 2021. PMID: 34227665 Free PMC article. Review.
-
Identification of a dCache-type chemoreceptor in Campylobacter jejuni that specifically mediates chemotaxis towards methyl pyruvate.Front Microbiol. 2024 May 9;15:1400284. doi: 10.3389/fmicb.2024.1400284. eCollection 2024. Front Microbiol. 2024. PMID: 38784811 Free PMC article.
-
Nitrate- and Nitrite-Sensing Histidine Kinases: Function, Structure, and Natural Diversity.Int J Mol Sci. 2021 May 31;22(11):5933. doi: 10.3390/ijms22115933. Int J Mol Sci. 2021. PMID: 34072989 Free PMC article. Review.
-
The Synthesis and Assembly of a Truncated Cyanophage Genome and Its Expression in a Heterogenous Host.Life (Basel). 2022 Aug 15;12(8):1234. doi: 10.3390/life12081234. Life (Basel). 2022. PMID: 36013413 Free PMC article.
-
Chemotaxis of Pseudomonas putida F1 to Alcohols Is Mediated by the Carboxylic Acid Receptor McfP.Appl Environ Microbiol. 2019 Oct 30;85(22):e01625-19. doi: 10.1128/AEM.01625-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31471307 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

