GH30-7 Endoxylanase C from the Filamentous Fungus Talaromyces cellulolyticus
- PMID: 31492671
- PMCID: PMC6821971
- DOI: 10.1128/AEM.01442-19
GH30-7 Endoxylanase C from the Filamentous Fungus Talaromyces cellulolyticus
Abstract
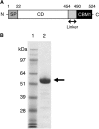
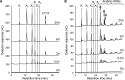
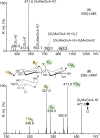
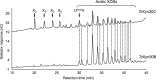
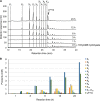
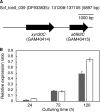
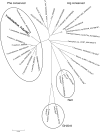
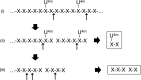
Glycoside hydrolase family 30 subfamily 7 (GH30-7) enzymes include various types of xylanases, such as glucuronoxylanase, endoxylanase, xylobiohydrolase, and reducing-end xylose-releasing exoxylanase. Here, we characterized the mode of action and gene expression of the GH30-7 endoxylanase from the cellulolytic fungus Talaromyces cellulolyticus (TcXyn30C). TcXyn30C has a modular structure consisting of a GH30-7 catalytic domain and a C-terminal cellulose binding module 1, whose cellulose-binding ability has been confirmed. Sequence alignment of GH30-7 xylanases exhibited that TcXyn30C has a conserved Phe residue at the position corresponding to a conserved Arg residue in GH30-7 glucuronoxylanases, which is required for the recognition of the 4-O-methyl-α-d-glucuronic acid (MeGlcA) substituent. TcXyn30C degraded both glucuronoxylan and arabinoxylan with similar kinetic constants and mainly produced linear xylooligosaccharides (XOSs) with 2 to 3 degrees of polymerization, in an endo manner. Notably, the hydrolysis of glucuronoxylan caused an accumulation of 22-(MeGlcA)-xylobiose (U4m2X). The production of this acidic XOS is likely to proceed via multistep reactions by putative glucuronoxylanase activity that produces 22-(MeGlcA)-XOSs (X n U4m2X, n ≥ 0) in the initial stages of the hydrolysis and by specific release of U4m2X from a mixture containing X n U4m2X. Our results suggest that the unique endoxylanase activity of TcXyn30C may be applicable to the production of linear and acidic XOSs. The gene xyn30C was located adjacent to the putative GH62 arabinofuranosidase gene (abf62C) in the T. cellulolyticus genome. The expression of both genes was induced by cellulose. The results suggest that TcXyn30C may be involved in xylan removal in the hydrolysis of lignocellulose by the T. cellulolyticus cellulolytic system.IMPORTANCE Xylooligosaccharides (XOSs), which are composed of xylose units with a β-1,4 linkage, have recently gained interest as prebiotics in the food and feed industry. Apart from linear XOSs, branched XOSs decorated with a substituent such as methyl glucuronic acid and arabinose also have potential applications. Endoxylanase is a promising tool in producing XOSs from xylan. The structural variety of XOSs generated depends on the substrate specificity of the enzyme as well as the distribution of the substituents in xylan. Thus, the exploration of endoxylanases with novel specificities is expected to be useful in the provision of a series of XOSs. In this study, the endoxylanase TcXyn30C from Talaromyces cellulolyticus was characterized as a unique glycoside hydrolase belonging to the family GH30-7, which specifically releases 22-(4-O-methyl-α-d-glucuronosyl)-xylobiose from hardwood xylan. This study provides new insights into the production of linear and branched XOSs by GH30-7 endoxylanase.
Keywords: Talaromyces cellulolyticus; endoxylanase; glycoside hydrolase family 30; lignocellulose; xylan; xylooligosaccharide.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Samanta AK, Jayapal N, Jayaram C, Roy S, Kolte AP, Senani S, Sridhar M. 2015. Xylooligosaccharides as prebiotics from agricultural by-products: production and applications. Bioact Carbohydr Diet Fibre 5:62–71. doi: 10.1016/j.bcdf.2014.12.003. - DOI
-
- Moure A, Gullón P, Domínguez H, Parajó JC. 2006. Advances in the manufacture, purification and applications of xylo-oligosaccharides as food additives and nutraceuticals. Process Biochem 41:1913–1923. doi: 10.1016/j.procbio.2006.05.011. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

