Comprehensive Analysis of the Lysine Succinylome and Protein Co-modifications in Developing Rice Seeds
- PMID: 31492684
- PMCID: PMC6885699
- DOI: 10.1074/mcp.RA119.001426
Comprehensive Analysis of the Lysine Succinylome and Protein Co-modifications in Developing Rice Seeds
Abstract
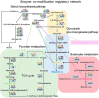
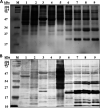
Lysine succinylation has been recognized as a post-translational modification (PTM) in recent years. It is plausible that succinylation may have a vaster functional impact than acetylation because of bulkier structural changes and more significant charge differences on the modified lysine residue. Currently, however, the quantity and identity of succinylated proteins and their corresponding functions in cereal plants remain largely unknown. In this study, we estimated the native succinylation occupancy on lysine was between 2% to 10% in developing rice seeds. Eight hundred fifty-four lysine succinylation sites on 347 proteins have been identified by a thorough investigation in developing rice seeds. Six motifs were revealed as preferred amino acid sequence arrangements for succinylation sites, and a noteworthy motif preference was identified in proteins associated with different biological processes, molecular functions, pathways, and domains. Remarkably, heavy succinylation was detected on major seed storage proteins, in conjunction with critical enzymes involved in central carbon metabolism and starch biosynthetic pathways for rice seed development. Meanwhile, our results showed that the modification pattern of in vitro nonenzymatically succinylated proteins was different from those of the proteins isolated from cells in Western blots, suggesting that succinylation is not generated via nonenzymatic reaction in the cells, at least not completely. Using the acylation data obtained from the same rice tissue, we mapped many sites harboring lysine succinylation, acetylation, malonylation, crotonylation, and 2-hydroxisobutyrylation in rice seed proteins. A striking number of proteins with multiple modifications were shown to be involved in critical metabolic events. Given that these modification moieties are intermediate products of multiple cellular metabolic pathways, these targeted lysine residues may mediate the crosstalk between different metabolic pathways via modifications by different moieties. Our study exhibits a platform for extensive investigation of molecular networks administrating cereal seed development and metabolism via PTMs.
Keywords: acetylation; lysine succinylation; mass spectrometry; plant biology; post-translational modifications; protein modification; rice; seeds; storage nutrient; succinylome.
© 2019 Meng et al.
Conflict of interest statement
The authors declare that they have no competing interests
Figures




Similar articles
-
Malonylome analysis in developing rice (Oryza sativa) seeds suggesting that protein lysine malonylation is well-conserved and overlaps with acetylation and succinylation substantially.J Proteomics. 2018 Jan 6;170:88-98. doi: 10.1016/j.jprot.2017.08.021. Epub 2017 Sep 4. J Proteomics. 2018. PMID: 28882676
-
Global Proteome Analyses of Lysine Acetylation and Succinylation Reveal the Widespread Involvement of both Modification in Metabolism in the Embryo of Germinating Rice Seed.J Proteome Res. 2016 Mar 4;15(3):879-90. doi: 10.1021/acs.jproteome.5b00805. Epub 2016 Jan 28. J Proteome Res. 2016. PMID: 26767346
-
Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation.Biochim Biophys Acta Proteins Proteom. 2018 Mar;1866(3):451-463. doi: 10.1016/j.bbapap.2017.12.001. Epub 2017 Dec 5. Biochim Biophys Acta Proteins Proteom. 2018. PMID: 29313810
-
Proteomics and Post-Translational Modifications of Starch Biosynthesis-Related Proteins in Developing Seeds of Rice.Int J Mol Sci. 2021 May 31;22(11):5901. doi: 10.3390/ijms22115901. Int J Mol Sci. 2021. PMID: 34072759 Free PMC article. Review.
-
Succinylation Links Metabolism to Protein Functions.Neurochem Res. 2019 Oct;44(10):2346-2359. doi: 10.1007/s11064-019-02780-x. Epub 2019 Mar 22. Neurochem Res. 2019. PMID: 30903449 Free PMC article. Review.
Cited by
-
The molecular basis of cereal grain proteostasis.Essays Biochem. 2022 Aug 5;66(2):243-253. doi: 10.1042/EBC20210041. Essays Biochem. 2022. PMID: 35818971 Free PMC article. Review.
-
Global analysis of lysine 2-hydroxyisobutyrylation in wheat root.Sci Rep. 2021 Mar 18;11(1):6327. doi: 10.1038/s41598-021-85879-y. Sci Rep. 2021. PMID: 33737719 Free PMC article.
-
Metabolite Profiling and Classification of Developing Styrax tonkinensis Kernels.Metabolites. 2020 Jan 1;10(1):21. doi: 10.3390/metabo10010021. Metabolites. 2020. PMID: 31906354 Free PMC article.
-
Protein post-translational modification by lysine succinylation: Biochemistry, biological implications, and therapeutic opportunities.Genes Dis. 2022 Apr 7;10(4):1242-1262. doi: 10.1016/j.gendis.2022.03.009. eCollection 2023 Jul. Genes Dis. 2022. PMID: 37397549 Free PMC article. Review.
-
Developmental Differences between Anthers of Diploid and Autotetraploid Rice at Meiosis.Plants (Basel). 2022 Jun 22;11(13):1647. doi: 10.3390/plants11131647. Plants (Basel). 2022. PMID: 35807599 Free PMC article.
References
-
- Weinert, B. T., Schölz, C., Wagner, S. A., Iesmantavicius, V., Su, D., Daniel, J. A., and Choudhary, C. (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

