MicroRNA-455-3p mediates GATA3 tumor suppression in mammary epithelial cells by inhibiting TGF-β signaling
- PMID: 31492753
- PMCID: PMC6816095
- DOI: 10.1074/jbc.RA119.010800
MicroRNA-455-3p mediates GATA3 tumor suppression in mammary epithelial cells by inhibiting TGF-β signaling
Abstract
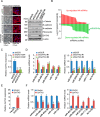
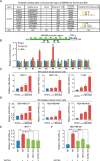
GATA3 is a basic and essential transcription factor that regulates many pathophysiological processes and is required for the development of mammary luminal epithelial cells. Loss-of-function GATA3 alterations in breast cancer are associated with poor prognosis. Here, we sought to understand the tumor-suppressive functions GATA3 normally performs. We discovered a role for GATA3 in suppressing epithelial-to-mesenchymal transition (EMT) in breast cancer by activating miR-455-3p expression. Enforced expression of miR-455-3p alone partially prevented EMT induced by transforming growth factor β (TGF-β) both in cells and tumor xenografts by directly inhibiting key components of TGF-β signaling. Pathway and biochemical analyses showed that one miRNA-455-3p target, the TGF-β-induced protein ZEB1, recruits the Mi-2/nucleosome remodeling and deacetylase (NuRD) complex to the promotor region of miR-455 to strictly repress the GATA3-induced transcription of this microRNA. Considering that ZEB1 enhances TGF-β signaling, we delineated a double-feedback interaction between ZEB1 and miR-455-3p, in addition to the repressive effect of miR-455-3p on TGF-β signaling. Our study revealed that a feedback loop between these two axes, specifically GATA3-induced miR-455-3p expression, could repress ZEB1 and its recruitment of NuRD (MTA1) to suppress miR-455, which ultimately regulates TGF-β signaling. In conclusion, we identified that miR-455-3p plays a pivotal role in inhibiting the EMT and TGF-β signaling pathway and maintaining cell differentiation. This forms the basis of that miR-455-3p might be a promising therapeutic intervention for breast cancer.
Keywords: GATA transcription factor; GATA3; HDAC2; Smad2; ZEB1; breast cancer; epithelial-mesenchymal transition (EMT); gene regulation; miR-455-3p; microRNA (miRNA); nucleosome remodeling deacetylase (NuRD); transforming growth factor beta (TGF-beta).
© 2019 Zeng et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





Similar articles
-
Suppression of breast cancer metastatic behavior by microRNAs targeting EMT transcription factors. A relevant participation of miR-196a-5p and miR-22-3p in ZEB1 expression.Breast Cancer Res Treat. 2025 Jul;212(2):277-290. doi: 10.1007/s10549-025-07723-5. Epub 2025 May 18. Breast Cancer Res Treat. 2025. PMID: 40382762
-
Copy number gain of ZEB1 mediates a double-negative feedback loop with miR-33a-5p that regulates EMT and bone metastasis of prostate cancer dependent on TGF-β signaling.Theranostics. 2019 Aug 14;9(21):6063-6079. doi: 10.7150/thno.36735. eCollection 2019. Theranostics. 2019. PMID: 31534537 Free PMC article.
-
miR-190 suppresses breast cancer metastasis by regulation of TGF-β-induced epithelial-mesenchymal transition.Mol Cancer. 2018 Mar 6;17(1):70. doi: 10.1186/s12943-018-0818-9. Mol Cancer. 2018. PMID: 29510731 Free PMC article.
-
miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance.J Mol Med (Berl). 2016 Jun;94(6):629-44. doi: 10.1007/s00109-016-1420-5. Epub 2016 Apr 20. J Mol Med (Berl). 2016. PMID: 27094812 Review.
-
Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer.J Transl Med. 2020 Feb 3;18(1):51. doi: 10.1186/s12967-020-02240-z. J Transl Med. 2020. PMID: 32014049 Free PMC article. Review.
Cited by
-
Regulation of breast cancer metastasis signaling by miRNAs.Cancer Metastasis Rev. 2020 Sep;39(3):837-886. doi: 10.1007/s10555-020-09905-7. Cancer Metastasis Rev. 2020. PMID: 32577859 Free PMC article. Review.
-
Long non-coding RNA LINC01116 is activated by EGR1 and facilitates lung adenocarcinoma oncogenicity via targeting miR-744-5p/CDCA4 axis.Cancer Cell Int. 2021 Jun 5;21(1):292. doi: 10.1186/s12935-021-01994-w. Cancer Cell Int. 2021. PMID: 34090440 Free PMC article.
-
MicroRNA-455-3P as a peripheral biomarker and therapeutic target for mild cognitive impairment and Alzheimer's disease.Ageing Res Rev. 2024 Sep;100:102459. doi: 10.1016/j.arr.2024.102459. Epub 2024 Aug 15. Ageing Res Rev. 2024. PMID: 39153602 Free PMC article. Review.
-
Regulation of gene expression by miRNA-455-3p, upregulated in the conjunctival epithelium of patients with Stevens-Johnson syndrome in the chronic stage.Sci Rep. 2020 Oct 14;10(1):17239. doi: 10.1038/s41598-020-74211-9. Sci Rep. 2020. PMID: 33057072 Free PMC article.
-
Long non-coding RNA FALEC promotes colorectal cancer progression via regulating miR-2116-3p-targeted PIWIL1.Cancer Biol Ther. 2020 Nov 1;21(11):1025-1032. doi: 10.1080/15384047.2020.1824514. Epub 2020 Oct 19. Cancer Biol Ther. 2020. PMID: 33073675 Free PMC article.
References
-
- Asselin-Labat M. L., Sutherland K. D., Barker H., Thomas R., Shackleton M., Forrest N. C., Hartley L., Robb L., Grosveld F. G., van der Wees J., Lindeman G. J., and Visvader J. E. (2007) Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 10.1038/ncb1530 - DOI - PubMed
-
- Usary J., Llaca V., Karaca G., Presswala S., Karaca M., He X., Langerød A., Kåresen R., Oh D. S., Dressler L. G., Lønning P. E., Strausberg R. L., Chanock S., Børresen-Dale A. L., and Perou C. M. (2004) Mutation of GATA3 in human breast tumors. Oncogene 23, 7669–7678 10.1038/sj.onc.1207966 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

