Odor-Specific Deactivation Defects in a Drosophila Odorant-Binding Protein Mutant
- PMID: 31492805
- PMCID: PMC6827369
- DOI: 10.1534/genetics.119.302629
Odor-Specific Deactivation Defects in a Drosophila Odorant-Binding Protein Mutant
Abstract
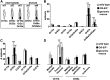
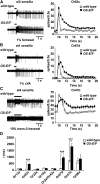
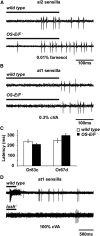
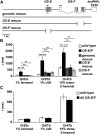
Insect odorant-binding proteins (OBPs) are a large, diverse group of low-molecular weight proteins secreted into the fluid bathing olfactory and gustatory neuron dendrites. The best-characterized OBP, LUSH (OBP76a) enhances pheromone sensitivity enabling detection of physiological levels of the male-specific pheromone, 11-cis vaccenyl acetate. The role of the other OBPs encoded in the Drosophila genome is largely unknown. Here, using clustered regularly interspaced short palindromic repeats/Cas9, we generated and characterized the loss-of-function phenotype for two genes encoding homologous OBPs, OS-E (OBP83b) and OS-F (OBP83a). Instead of activation defects, these extracellular proteins are required for normal deactivation of odorant responses to a subset of odorants. Remarkably, odorants detected by the same odorant receptor are differentially affected by the loss of the OBPs, revealing an odorant-specific role in deactivation kinetics. In stark contrast to lush mutants, the OS-E/F mutants have normal activation kinetics to the affected odorants, even at low stimulus concentrations, suggesting that these OBPs are not competing for these ligands with the odorant receptors. We also show that OS-E and OS-F are functionally redundant as either is sufficient to revert the mutant phenotype in transgenic rescue experiments. These findings expand our understanding of the roles of OBPs to include the deactivation of odorant responses.
Keywords: olfaction; olfactory; perireceptor.
Copyright © 2019 by the Genetics Society of America.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

