RNA editing alterations define manifestation of prion diseases
- PMID: 31492812
- PMCID: PMC6765247
- DOI: 10.1073/pnas.1803521116
RNA editing alterations define manifestation of prion diseases
Abstract
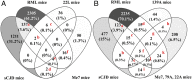
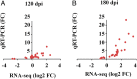
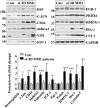
Prion diseases are fatal neurodegenerative disorders caused by misfolding of the normal prion protein into an infectious cellular pathogen. Clinically characterized by rapidly progressive dementia and accounting for 85% of human prion disease cases, sporadic Creutzfeldt-Jakob disease (sCJD) is the prevalent human prion disease. Although sCJD neuropathological hallmarks are well-known, associated molecular alterations are elusive due to rapid progression and absence of preclinical stages. To investigate transcriptome alterations during disease progression, we utilized tg340-PRNP129MM mice infected with postmortem material from sCJD patients of the most susceptible genotype (MM1 subtype), a sCJD model that faithfully recapitulates the molecular and pathological alterations of the human disease. Here we report that transcriptomic analyses from brain cortex in the context of disease progression, reveal epitranscriptomic alterations (specifically altered RNA edited pathway profiles, eg., ER stress, lysosome) that are characteristic and possibly protective mainly for preclinical and clinical disease stages. Our results implicate regulatory epitranscriptomic mechanisms in prion disease neuropathogenesis, whereby RNA-editing targets in a humanized sCJD mouse model were confirmed in pathological human autopsy material.
Keywords: ER-stress; RNA editing; RNA-sequencing; prion diseases; sporadic Creutzfeldt–Jakob disease.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

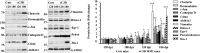
Similar articles
-
Regional and subtype-dependent miRNA signatures in sporadic Creutzfeldt-Jakob disease are accompanied by alterations in miRNA silencing machinery and biogenesis.PLoS Pathog. 2018 Jan 22;14(1):e1006802. doi: 10.1371/journal.ppat.1006802. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29357384 Free PMC article.
-
Subtype and regional regulation of prion biomarkers in sporadic Creutzfeldt-Jakob disease.Neuropathol Appl Neurobiol. 2015 Aug;41(5):631-45. doi: 10.1111/nan.12175. Epub 2015 Apr 30. Neuropathol Appl Neurobiol. 2015. PMID: 25134744
-
Prion Strain Characterization of a Novel Subtype of Creutzfeldt-Jakob Disease.J Virol. 2017 May 12;91(11):e02390-16. doi: 10.1128/JVI.02390-16. Print 2017 Jun 1. J Virol. 2017. PMID: 28298604 Free PMC article.
-
Genetic risk factors for Creutzfeldt-Jakob disease.Neurobiol Dis. 2020 Aug;142:104973. doi: 10.1016/j.nbd.2020.104973. Epub 2020 Jun 18. Neurobiol Dis. 2020. PMID: 32565065 Review.
-
Genetic and Transcriptomic Profiles of Inflammation in Neurodegenerative Diseases: Alzheimer, Parkinson, Creutzfeldt-Jakob and Tauopathies.Int J Mol Sci. 2016 Feb 4;17(2):206. doi: 10.3390/ijms17020206. Int J Mol Sci. 2016. PMID: 26861289 Free PMC article. Review.
Cited by
-
Genome-wide transcriptomics identifies an early preclinical signature of prion infection.PLoS Pathog. 2020 Jun 29;16(6):e1008653. doi: 10.1371/journal.ppat.1008653. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32598380 Free PMC article.
-
Exploring Additional Valuable Information From Single-Cell RNA-Seq Data.Front Cell Dev Biol. 2020 Dec 1;8:593007. doi: 10.3389/fcell.2020.593007. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33335900 Free PMC article. Review.
-
Prion diseases disrupt glutamate/glutamine metabolism in skeletal muscle.PLoS Pathog. 2024 Sep 11;20(9):e1012552. doi: 10.1371/journal.ppat.1012552. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39259763 Free PMC article.
-
Increased Alu RNA processing in Alzheimer brains is linked to gene expression changes.EMBO Rep. 2021 May 5;22(5):e52255. doi: 10.15252/embr.202052255. Epub 2021 Mar 1. EMBO Rep. 2021. PMID: 33645898 Free PMC article.
-
Genomic, transcriptomic and RNA editing analysis of human MM1 and VV2 sporadic Creutzfeldt-Jakob disease.Acta Neuropathol Commun. 2022 Dec 14;10(1):181. doi: 10.1186/s40478-022-01483-9. Acta Neuropathol Commun. 2022. PMID: 36517866 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

