Revealing the metabolic capacity of Streblomastix strix and its bacterial symbionts using single-cell metagenomics
- PMID: 31492817
- PMCID: PMC6765251
- DOI: 10.1073/pnas.1910793116
Revealing the metabolic capacity of Streblomastix strix and its bacterial symbionts using single-cell metagenomics
Abstract
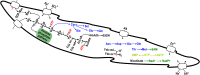
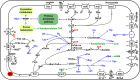
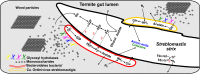
Lower termites harbor in their hindgut complex microbial communities that are involved in the digestion of cellulose. Among these are protists, which are usually associated with specific bacterial symbionts found on their surface or inside their cells. While these form the foundations of a classic system in symbiosis research, we still know little about the functional basis for most of these relationships. Here, we describe the complex functional relationship between one protist, the oxymonad Streblomastix strix, and its ectosymbiotic bacterial community using single-cell genomics. We generated partial assemblies of the host S. strix genome and Candidatus Ordinivivax streblomastigis, as well as a complex metagenome assembly of at least 8 other Bacteroidetes bacteria confirmed by ribosomal (r)RNA fluorescence in situ hybridization (FISH) to be associated with S. strix. Our data suggest that S. strix is probably not involved in the cellulose digestion, but the bacterial community on its surface secretes a complex array of glycosyl hydrolases, providing them with the ability to degrade cellulose to monomers and fueling the metabolism of S. strix In addition, some of the bacteria can fix nitrogen and can theoretically provide S. strix with essential amino acids and cofactors, which the protist cannot synthesize. On the contrary, most of the bacterial symbionts lack the essential glycolytic enzyme enolase, which may be overcome by the exchange of intermediates with S. strix This study demonstrates the value of the combined single-cell (meta)genomic and FISH approach for studies of complicated symbiotic systems.
Keywords: Bacteroidetes; Streblomastix; ectosymbionts; oxymonads; termite.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Brune A., Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180 (2014). - PubMed
-
- Brune A., “Methanogens in the digestive tract of termites” in (Endo)Symbiotic Methanogenic Archaea, Hackstein J. H. P., Ed. (Springer Berlin Heidelberg, 2018), pp. 81–101.
-
- Strassert J. F. H., Mikaelyan A., Woyke T., Brune A., Genome analysis of ‘Candidatus Ancillula trichonymphae’, first representative of a deep-branching clade of Bifidobacteriales, strengthens evidence for convergent evolution in flagellate endosymbionts. Environ. Microbiol. Rep. 8, 865–873 (2016). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

