CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation
- PMID: 31492834
- PMCID: PMC6731291
- DOI: 10.1038/s41467-019-12028-5
CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation
Abstract
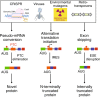
The introduction of insertion-deletions (INDELs) by non-homologous end-joining (NHEJ) pathway underlies the mechanistic basis of CRISPR-Cas9-directed genome editing. Selective gene ablation using CRISPR-Cas9 is achieved by installation of a premature termination codon (PTC) from a frameshift-inducing INDEL that elicits nonsense-mediated decay (NMD) of the mutant mRNA. Here, by examining the mRNA and protein products of CRISPR targeted genes in a cell line panel with presumed gene knockouts, we detect the production of foreign mRNAs or proteins in ~50% of the cell lines. We demonstrate that these aberrant protein products stem from the introduction of INDELs that promote internal ribosomal entry, convert pseudo-mRNAs (alternatively spliced mRNAs with a PTC) into protein encoding molecules, or induce exon skipping by disruption of exon splicing enhancers (ESEs). Our results reveal challenges to manipulating gene expression outcomes using INDEL-based mutagenesis and strategies useful in mitigating their impact on intended genome-editing outcomes.
Conflict of interest statement
The authors L.L., R.T., T.H.H., Y.Y., J.T.P. and Q.B. are named inventors on a patent (under consideration: 16/003683) applied for by University of Texas Southwestern Medical Center, Dallas, on behalf of the inventors that covers strategies for inducing or avoiding exon skipping using CRISPR — specifically the CRISPinator algorithm and targeting exon splicing enhancers using INDELs to induce exon skipping with a single CRISPR guide strategy. The remaining authors declare no competing interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

