Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection
- PMID: 31492859
- PMCID: PMC6731267
- DOI: 10.1038/s41467-019-12064-1
Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection
Abstract
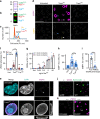
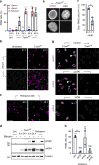
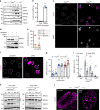
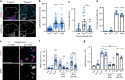
Salmonella Typhi activates the host DNA damage response through the typhoid toxin, facilitating typhoid symptoms and chronic infections. Here we reveal a non-canonical DNA damage response, which we call RING (response induced by a genotoxin), characterized by accumulation of phosphorylated histone H2AX (γH2AX) at the nuclear periphery. RING is the result of persistent DNA damage mediated by toxin nuclease activity and is characterized by hyperphosphorylation of RPA, a sensor of single-stranded DNA (ssDNA) and DNA replication stress. The toxin overloads the RPA pathway with ssDNA substrate, causing RPA exhaustion and senescence. Senescence is also induced by canonical γΗ2ΑΧ foci revealing distinct mechanisms. Senescence is transmitted to non-intoxicated bystander cells by an unidentified senescence-associated secreted factor that enhances Salmonella infections. Thus, our work uncovers a mechanism by which genotoxic Salmonella exhausts the RPA response by inducing ssDNA formation, driving host cell senescence and facilitating infection.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

