In vitro role of Rad54 in Rad51-ssDNA filament-dependent homology search and synaptic complexes formation
- PMID: 31492866
- PMCID: PMC6731316
- DOI: 10.1038/s41467-019-12082-z
In vitro role of Rad54 in Rad51-ssDNA filament-dependent homology search and synaptic complexes formation
Abstract
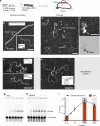
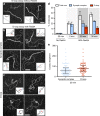
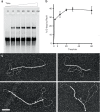
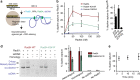
Homologous recombination (HR) uses a homologous template to accurately repair DNA double-strand breaks and stalled replication forks to maintain genome stability. During homology search, Rad51 nucleoprotein filaments probe and interact with dsDNA, forming the synaptic complex that is stabilized on a homologous sequence. Strand intertwining leads to the formation of a displacement-loop (D-loop). In yeast, Rad54 is essential for HR in vivo and required for D-loop formation in vitro, but its exact role remains to be fully elucidated. Using electron microscopy to visualize the DNA-protein complexes, here we find that Rad54 is crucial for Rad51-mediated synaptic complex formation and homology search. The Rad54-K341R ATPase-deficient mutant protein promotes formation of synaptic complexes but not D-loops and leads to the accumulation of stable heterologous associations, suggesting that the Rad54 ATPase is involved in preventing non-productive intermediates. We propose that Rad51/Rad54 form a functional unit operating in homology search, synaptic complex and D-loop formation.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

