Metagenomic and chemical characterization of soil cobalamin production
- PMID: 31492962
- PMCID: PMC6908642
- DOI: 10.1038/s41396-019-0502-0
Metagenomic and chemical characterization of soil cobalamin production
Abstract
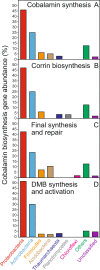
Cobalamin (vitamin B12) is an essential enzyme cofactor for most branches of life. Despite the potential importance of this cofactor for soil microbial communities, the producers and consumers of cobalamin in terrestrial environments are still unknown. Here we provide the first metagenome-based assessment of soil cobalamin-producing bacteria and archaea, quantifying and classifying genes encoding proteins for cobalamin biosynthesis, transport, remodeling, and dependency in 155 soil metagenomes with profile hidden Markov models. We also measured several forms of cobalamin (CN-, Me-, OH-, Ado-B12) and the cobalamin lower ligand (5,6-dimethylbenzimidazole; DMB) in 40 diverse soil samples. Metagenomic analysis revealed that less than 10% of soil bacteria and archaea encode the genetic potential for de novo synthesis of this important enzyme cofactor. Predominant soil cobalamin producers were associated with the Proteobacteria, Actinobacteria, Firmicutes, Nitrospirae, and Thaumarchaeota. In contrast, a much larger proportion of abundant soil genera lacked cobalamin synthesis genes and instead were associated with gene sequences encoding cobalamin transport and cobalamin-dependent enzymes. The enrichment of DMB and corresponding DMB synthesis genes, relative to corrin ring synthesis genes, suggests an important role for cobalamin remodelers in terrestrial habitats. Together, our results indicate that microbial cobalamin production and repair serve as keystone functions that are significantly correlated with microbial community size, diversity, and biogeochemistry of terrestrial ecosystems.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures