The N-terminal D1 domain of Treponema pallidum flagellin binding to TLR5 is required but not sufficient in activation of TLR5
- PMID: 31493340
- PMCID: PMC6815820
- DOI: 10.1111/jcmm.14617
The N-terminal D1 domain of Treponema pallidum flagellin binding to TLR5 is required but not sufficient in activation of TLR5
Abstract
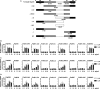
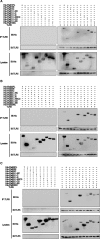
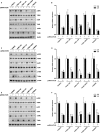
Syphilis is a chronic bacterial infection caused by Treponema pallidum (T pallidum) and the pathogenesis that T pallidum infection induces immunopathological damages in skin and other tissues remains unclear. We have previously reported that recombinant flagellins of T pallidum can elicit IL-6 and IL-8 transcriptions via TLR5 pathway. To identify the domains which induced the pro-inflammatory activity and the importance of the interactions between TLR5 and domains, homology-based modelling and comparative structural analyses revealed that Tpflagellins can combine with TLR5 directly. Deletion mutations showed that the ND1 domain binding to TLR5 is required but not sufficient in TLR5 activation. Moreover, site-directed mutagenesis analysis indicated that the arginine residue (Tpflagellins R89) of the ND1 domain and its adjacent residues (Tpflagellins L93 and E113) constitute a hot spot that elicits IL-6, IL-8 transcriptions and TLR5 activation, and affects the binding of Tpflagellins to TLR5. Taken together, these results give insight into the pathogenesis of T pallidum and may contribute to the future design of Tpflagellins-based therapeutics and syphilis vaccine.
Keywords: Treponema pallidum; N-terminal D1 domain; TLR5 signalling; flagellin; inflammation.
© 2019 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

