Transcriptomic Dissection of Hepatocyte Heterogeneity: Linking Ploidy, Zonation, and Stem/Progenitor Cell Characteristics
- PMID: 31493546
- PMCID: PMC6909008
- DOI: 10.1016/j.jcmgh.2019.08.011
Transcriptomic Dissection of Hepatocyte Heterogeneity: Linking Ploidy, Zonation, and Stem/Progenitor Cell Characteristics
Abstract
Background & aims: There is a long-standing debate regarding the biological significance of polyploidy in hepatocytes. Recent studies have provided increasing evidence that hepatocytes with different ploidy statuses behave differently in a context-dependent manner (eg, susceptibility to oncogenesis, regenerative ability after injury, and in vitro proliferative capacity). However, their overall transcriptomic differences in a physiological context is not known.
Methods: By using microarray transcriptome analysis, we investigated the heterogeneity of hepatocyte populations with different ploidy statuses. Moreover, by using single-cell quantitative reverse-transcription polymerase chain reaction (scPCR) analysis, we investigated the intrapopulational transcriptome heterogeneity of 2c and 4c hepatocytes.
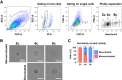
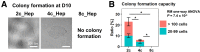
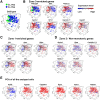
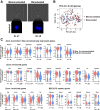
Results: Microarray analysis showed that cell cycle-related genes were enriched in 8c hepatocytes, which is in line with the established notion that polyploidy is formed via cell division failure. Surprisingly, in contrast to the general consensus that 2c hepatocytes reside in the periportal region, in our bulk transcriptome and scPCR analyses, the 2c hepatocytes consistently showed pericentral hepatocyte-enriched characteristics. In addition, scPCR analysis identified a subpopulation within the 2c hepatocytes that co-express the liver progenitor cell markers Axin2, Prom1, and Lgr5, implying the potential biological relevance of this subpopulation.
Conclusions: This study provides new insights into hepatocyte heterogeneity, namely 2c hepatocytes are preferentially localized to the pericentral region, and a subpopulation of 2c hepatocytes show liver progenitor cell-like features in terms of liver progenitor cell marker expression (Axin2, Prom1, and Lgr5).
Keywords: Hepatocyte; Ploidy; Single-Cell PCR; Transcriptome; Zonation.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures








Comment in
-
Single-Cell and Bulk Transcriptome Profiling Reveals Unique Features of Diploid and Polyploid Hepatocytes.Cell Mol Gastroenterol Hepatol. 2020;9(1):193-194. doi: 10.1016/j.jcmgh.2019.09.008. Epub 2019 Oct 22. Cell Mol Gastroenterol Hepatol. 2020. PMID: 31654613 Free PMC article. No abstract available.
References
-
- Gentric G., Celton-Morizur S., Desdouets C. Polyploidy and liver proliferation. Clin Res Hepatol Gastroenterol. 2012;36:29–34. - PubMed
-
- Gentric G., Desdouets C. Polyploidization in liver tissue. Am J Pathol. 2014;184:322–331. - PubMed
-
- Pandit S.K., Westendorp B., De Bruin A. Physiological significance of polyploidization in mammalian cells. Trends Cell Biol. 2013;23:556–566. - PubMed
-
- Duncan A.W. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. 2013;24:347–356. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

