Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation
- PMID: 31494034
- PMCID: PMC6838673
- DOI: 10.1016/j.molcel.2019.07.038
Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation
Abstract
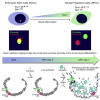
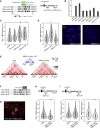
Enhancers can regulate the promoters of their target genes over very large genomic distances. It is widely assumed that mechanisms of enhancer action involve the reorganization of three-dimensional chromatin architecture, but this is poorly understood. The predominant model involves physical enhancer-promoter interaction by looping out the intervening chromatin. However, studying the enhancer-driven activation of the Sonic hedgehog gene (Shh), we have identified a change in chromosome conformation that is incompatible with this simple looping model. Using super-resolution 3D-FISH and chromosome conformation capture, we observe a decreased spatial proximity between Shh and its enhancers during the differentiation of embryonic stem cells to neural progenitors. We show that this can be recapitulated by synthetic enhancer activation, is impeded by chromatin-bound proteins located between the enhancer and the promoter, and appears to involve the catalytic activity of poly (ADP-ribose) polymerase. Our data suggest that models of enhancer-promoter communication need to encompass chromatin conformations other than looping.
Keywords: PARP; Sonic hedgehog; TAL-effector; chromatin looping; enhancer; poly(ADP-ribosyl)ation.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

