High-sensitivity microsatellite instability assessment for the detection of mismatch repair defects in normal tissue of biallelic germline mismatch repair mutation carriers
- PMID: 31494577
- PMCID: PMC7146943
- DOI: 10.1136/jmedgenet-2019-106272
High-sensitivity microsatellite instability assessment for the detection of mismatch repair defects in normal tissue of biallelic germline mismatch repair mutation carriers
Abstract
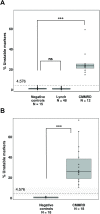
Introduction: Lynch syndrome (LS) and constitutional mismatch repair deficiency (CMMRD) are hereditary cancer syndromes associated with mismatch repair (MMR) deficiency. Tumours show microsatellite instability (MSI), also reported at low levels in non-neoplastic tissues. Our aim was to evaluate the performance of high-sensitivity MSI (hs-MSI) assessment for the identification of LS and CMMRD in non-neoplastic tissues.
Materials and methods: Blood DNA samples from 131 individuals were grouped into three cohorts: baseline (22 controls), training (11 CMMRD, 48 LS and 15 controls) and validation (18 CMMRD and 18 controls). Custom next generation sequencing panel and bioinformatics pipeline were used to detect insertions and deletions in microsatellite markers. An hs-MSI score was calculated representing the percentage of unstable markers.
Results: The hs-MSI score was significantly higher in CMMRD blood samples when compared with controls in the training cohort (p<0.001). This finding was confirmed in the validation set, reaching 100% specificity and sensitivity. Higher hs-MSI scores were detected in biallelic MSH2 carriers (n=5) compared with MSH6 carriers (n=15). The hs-MSI analysis did not detect a difference between LS and control blood samples (p=0.564).
Conclusions: The hs-MSI approach is a valuable tool for CMMRD diagnosis, especially in suspected patients harbouring MMR variants of unknown significance or non-detected biallelic germline mutations.
Keywords: constitutional mismatch repair deficiency; highly sensitive methodologies; lynch syndrome; microsatellite instability; next generation sequencing.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
-
- Bakry D, Aronson M, Durno C, Rimawi H, Farah R, Alharbi QK, Alharbi M, Shamvil A, Ben-Shachar S, Mistry M, Constantini S, Dvir R, Qaddoumi I, Gallinger S, Lerner-Ellis J, Pollett A, Stephens D, Kelies S, Chao E, Malkin D, Bouffet E, Hawkins C, Tabori U. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency Consortium. Eur J Cancer 2014;50:987–96. 10.1016/j.ejca.2013.12.005 - DOI - PubMed
-
- Wimmer K, Kratz CP, Vasen HFA, Caron O, Colas C, Entz-Werle N, Gerdes A-M, Goldberg Y, Ilencikova D, Muleris M, Duval A, Lavoine N, Ruiz-Ponte C, Slavc I, Burkhardt B, Brugieres L, EU-Consortium Care for CMMRD (C4CMMRD) . Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium 'care for CMMRD' (C4CMMRD). J Med Genet 2014;51:355–65. 10.1136/jmedgenet-2014-102284 - DOI - PubMed
-
- Li L, Hamel N, Baker K, McGuffin MJ, Couillard M, Gologan A, Marcus VA, Chodirker B, Chudley A, Stefanovici C, Durandy A, Hegele RA, Feng B-J, Goldgar DE, Zhu J, De Rosa M, Gruber SB, Wimmer K, Young B, Chong G, Tischkowitz MD, Foulkes WD. A homozygous PMS2 founder mutation with an attenuated constitutional mismatch repair deficiency phenotype. J Med Genet 2015;52:348–52. 10.1136/jmedgenet-2014-102934 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
