Validated inference of smoking habits from blood with a finite DNA methylation marker set
- PMID: 31494793
- PMCID: PMC6861351
- DOI: 10.1007/s10654-019-00555-w
Validated inference of smoking habits from blood with a finite DNA methylation marker set
Abstract
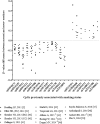
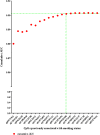
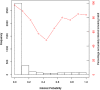
Inferring a person's smoking habit and history from blood is relevant for complementing or replacing self-reports in epidemiological and public health research, and for forensic applications. However, a finite DNA methylation marker set and a validated statistical model based on a large dataset are not yet available. Employing 14 epigenome-wide association studies for marker discovery, and using data from six population-based cohorts (N = 3764) for model building, we identified 13 CpGs most suitable for inferring smoking versus non-smoking status from blood with a cumulative Area Under the Curve (AUC) of 0.901. Internal fivefold cross-validation yielded an average AUC of 0.897 ± 0.137, while external model validation in an independent population-based cohort (N = 1608) achieved an AUC of 0.911. These 13 CpGs also provided accurate inference of current (average AUCcrossvalidation 0.925 ± 0.021, AUCexternalvalidation0.914), former (0.766 ± 0.023, 0.699) and never smoking (0.830 ± 0.019, 0.781) status, allowed inferring pack-years in current smokers (10 pack-years 0.800 ± 0.068, 0.796; 15 pack-years 0.767 ± 0.102, 0.752) and inferring smoking cessation time in former smokers (5 years 0.774 ± 0.024, 0.760; 10 years 0.766 ± 0.033, 0.764; 15 years 0.767 ± 0.020, 0.754). Model application to children revealed highly accurate inference of the true non-smoking status (6 years of age: accuracy 0.994, N = 355; 10 years: 0.994, N = 309), suggesting prenatal and passive smoking exposure having no impact on model applications in adults. The finite set of DNA methylation markers allow accurate inference of smoking habit, with comparable accuracy as plasma cotinine use, and smoking history from blood, which we envision becoming useful in epidemiology and public health research, and in medical and forensic applications.
Keywords: DNA methylation; Epidemiology; Epigenetics; Forensics; Smoking inference.
Conflict of interest statement
H.J. Grabe has received funding from Fresenius Medical Care and speaker’s honoraria as well as travel funds from Fresenius Medical Care, Neuraxpharm and Janssen-Cilag. Other than that, the authors declared no conflict of interest.
Figures
Similar articles
-
DNA methylation and smoking in Korean adults: epigenome-wide association study.Clin Epigenetics. 2016 Sep 22;8:103. doi: 10.1186/s13148-016-0266-6. eCollection 2016. Clin Epigenetics. 2016. PMID: 27688819 Free PMC article.
-
Associations of self-reported smoking, cotinine levels and epigenetic smoking indicators with oxidative stress among older adults: a population-based study.Eur J Epidemiol. 2017 May;32(5):443-456. doi: 10.1007/s10654-017-0248-9. Epub 2017 Apr 22. Eur J Epidemiol. 2017. PMID: 28434075
-
Self-reported smoking, serum cotinine, and blood DNA methylation.Environ Res. 2016 Apr;146:395-403. doi: 10.1016/j.envres.2016.01.026. Epub 2016 Jan 29. Environ Res. 2016. PMID: 26826776
-
Contrasting the effects of intra-uterine smoking and one-carbon micronutrient exposures on offspring DNA methylation.Epigenomics. 2017 Mar;9(3):351-367. doi: 10.2217/epi-2016-0135. Epub 2017 Feb 17. Epigenomics. 2017. PMID: 28234021 Free PMC article. Review.
-
DNA Methylation-Based Biomarkers of Environmental Exposures for Human Population Studies.Curr Environ Health Rep. 2020 Jun;7(2):121-128. doi: 10.1007/s40572-020-00269-2. Curr Environ Health Rep. 2020. PMID: 32062850 Review.
Cited by
-
Derivation and validation of an epigenetic frailty risk score in population-based cohorts of older adults.Nat Commun. 2022 Sep 7;13(1):5269. doi: 10.1038/s41467-022-32893-x. Nat Commun. 2022. PMID: 36071044 Free PMC article.
-
DNA methylation at AHRR as a master predictor of smoke exposure and a biomarker for sleep and exercise.Clin Epigenetics. 2024 Oct 18;16(1):147. doi: 10.1186/s13148-024-01757-0. Clin Epigenetics. 2024. PMID: 39425209 Free PMC article.
-
Epigenetic modelling of former, current and never smokers.Clin Epigenetics. 2021 Nov 17;13(1):206. doi: 10.1186/s13148-021-01191-6. Clin Epigenetics. 2021. PMID: 34789321 Free PMC article.
-
Objectives, design and main findings until 2020 from the Rotterdam Study.Eur J Epidemiol. 2020 May;35(5):483-517. doi: 10.1007/s10654-020-00640-5. Epub 2020 May 4. Eur J Epidemiol. 2020. PMID: 32367290 Free PMC article.
-
Epigenetic biomarkers for smoking cessation.Addict Neurosci. 2023 Jun;6:100079. doi: 10.1016/j.addicn.2023.100079. Epub 2023 Mar 1. Addict Neurosci. 2023. PMID: 37123087 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- NWO 184.021.007/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 529051014/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 016.136.361/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 184021007/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 050-060-810/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 633595/Horizon 2020 Framework Programme
- 733206/Horizon 2020 Framework Programme
- 696295/Horizon 2020 Framework Programme
- ERC-2014-CoG-648916/H2020 European Research Council
- 603288/FP7 Science in Society
- 602736/FP7 Science in Society
- R01HD068437/National Institute of Child Health and Human Development
- 01ZZ9603/Bundesministerium für Bildung und Forschung
- 01ZZ0103/Bundesministerium für Bildung und Forschung
- 01ZZ0403/Bundesministerium für Bildung und Forschung
- 03IS2061A/Bundesministerium für Bildung und Forschung
- 81X3400104/Deutsches Zentrum für Herz- Kreislaufforschung
- 940-35-034/Netherlands Organization for Scientific Research
- 98.901/Dutch Diabetes Research Foundation
- NWO-Groot 480-15-001/674/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 904-61-090/ZonMw
- 985-10-002/ZonMw
- 912-10-020/ZonMw
- 904-61-193/ZonMw
- 480-04-004/ZonMw
- 463-06-001/ZonMw
- 451-04-034/ZonMw
- 400-05-717/ZonMw
LinkOut - more resources
Full Text Sources

