Loss of SMPD4 Causes a Developmental Disorder Characterized by Microcephaly and Congenital Arthrogryposis
- PMID: 31495489
- PMCID: PMC6817560
- DOI: 10.1016/j.ajhg.2019.08.006
Loss of SMPD4 Causes a Developmental Disorder Characterized by Microcephaly and Congenital Arthrogryposis
Abstract
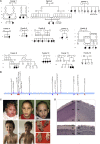
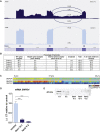
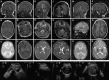
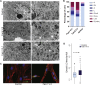
Sphingomyelinases generate ceramide from sphingomyelin as a second messenger in intracellular signaling pathways involved in cell proliferation, differentiation, or apoptosis. Children from 12 unrelated families presented with microcephaly, simplified gyral pattern of the cortex, hypomyelination, cerebellar hypoplasia, congenital arthrogryposis, and early fetal/postnatal demise. Genomic analysis revealed bi-allelic loss-of-function variants in SMPD4, coding for the neutral sphingomyelinase-3 (nSMase-3/SMPD4). Overexpression of human Myc-tagged SMPD4 showed localization both to the outer nuclear envelope and the ER and additionally revealed interactions with several nuclear pore complex proteins by proteomics analysis. Fibroblasts from affected individuals showed ER cisternae abnormalities, suspected for increased autophagy, and were more susceptible to apoptosis under stress conditions, while treatment with siSMPD4 caused delayed cell cycle progression. Our data show that SMPD4 links homeostasis of membrane sphingolipids to cell fate by regulating the cross-talk between the ER and the outer nuclear envelope, while its loss reveals a pathogenic mechanism in microcephaly.
Keywords: NET13; SMPD4; arthrogryposis; microcephaly; neutral-sphingomyelinase.
Copyright © 2019 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Bienias K., Fiedorowicz A., Sadowska A., Prokopiuk S., Car H. Regulation of sphingomyelin metabolism. Pharmacol. Rep. 2016;68:570–581. - PubMed
-
- Krut O., Wiegmann K., Kashkar H., Yazdanpanah B., Krönke M. Novel tumor necrosis factor-responsive mammalian neutral sphingomyelinase-3 is a C-tail-anchored protein. J. Biol. Chem. 2006;281:13784–13793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

