High-Throughput Single-Cell Sequencing with Linear Amplification
- PMID: 31495564
- PMCID: PMC6874760
- DOI: 10.1016/j.molcel.2019.08.002
High-Throughput Single-Cell Sequencing with Linear Amplification
Abstract
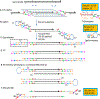
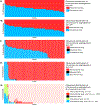
Conventional methods for single-cell genome sequencing are limited with respect to uniformity and throughput. Here, we describe sci-L3, a single-cell sequencing method that combines combinatorial indexing (sci-) and linear (L) amplification. The sci-L3 method adopts a 3-level (3) indexing scheme that minimizes amplification biases while enabling exponential gains in throughput. We demonstrate the generalizability of sci-L3 with proof-of-concept demonstrations of single-cell whole-genome sequencing (sci-L3-WGS), targeted sequencing (sci-L3-target-seq), and a co-assay of the genome and transcriptome (sci-L3-RNA/DNA). We apply sci-L3-WGS to profile the genomes of >10,000 sperm and sperm precursors from F1 hybrid mice, mapping 86,786 crossovers and characterizing rare chromosome mis-segregation events in meiosis, including instances of whole-genome equational chromosome segregation. We anticipate that sci-L3 assays can be applied to fully characterize recombination landscapes, to couple CRISPR perturbations and measurements of genome stability, and to other goals requiring high-throughput, high-coverage single-cell sequencing.
Keywords: DNA repair; chromosome segregation; double-strand break; homologous recombination; infertility; linear amplification; meiotic crossover; mouse; single-cell combinatorial indexing; single-cell sequencing.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
F.J.S. declares competing financial interests in the form of stock ownership and paid employment by Illumina. One or more embodiments of one or more patents and patent applications by the University of Washington and Illumina may encompass the methods, reagents, and data disclosed in this manuscript.
Figures




References
-
- Barrai S, Morozumi Y, Tanaka H, Montellier E, Govin J, de Dieuleveult M, Charbonnier G, Couté Y, Puthier D, Buchou T, et al. (2017). Histone Variant H2A.L.2 Guides Transition Protein-Dependent Protamine Assembly in Male Germ Cells. Mol. Cell 66, 89–101.e8. - PubMed
-
- Baudat F, Manova K, Yuen JP, Jasin M, and Keeney S (2000). Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spoil. Mol. Cell 6, 989–998. - PubMed
-
- Baudat F, Imai Y, and de Massy B (2013). Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet 14, 794–806. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

