tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer
- PMID: 31496739
- PMCID: PMC6702494
- DOI: 10.2147/OTT.S206861
tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer
Abstract
Background: High-grade serous ovarian cancer (HGSOC) is one of the most common ovarian epithelial malignancies. tRNA-derived fragments (tRFs) have been identified as novel potential biomarkers and targets for cancer therapy. Nevertheless, the influence of tRFs on HGSOC remains unknown. This study aimed to identify HGSOC-associated tRFs and to investigate the function and mechanism of key tRFs in SK-OV-3 ovarian cancer cells.
Methods: The tRF profiles in HGSOC patients and controls were investigated using small RNA sequencing. Differentially expressed tRFs were verified by real-time PCR, and a key tRF was evaluated in a function study.
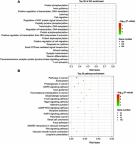
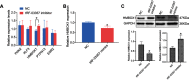
Results: A total of 27 tRFs were differentially expressed between HGSOC patients and controls. Differentially expressed tRFs were mainly involved in the functions of protein phosphorylation, transcription and cell migration and the pathway of cancer, and the MAPK and Wnt signaling pathways. Real-time PCR verified that tRF-03357 and tRF-03358 were significantly increased in the HGSOC serum samples and SK-OV-3 cells compared to their expression levels in the controls. Importantly, tRF-03357 promoted SK-OV-3 cell proliferation, migration and invasion. Moreover, tRF-03357 was predictively targeted, and significantly downregulated HMBOX1.
Conclusion: This study suggests that tRF-03357 might promote cell proliferation, migration and invasion, partly by modulating HMBOX1 in HGSOC.
Keywords: cell growth; high-grade serous ovarian cancer; invasion; migration; tRNA-derived fragments.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures



Similar articles
-
Differential Expression Profiles and Function Prediction of Transfer RNA-Derived Fragments in High-Grade Serous Ovarian Cancer.Biomed Res Int. 2021 Mar 30;2021:5594081. doi: 10.1155/2021/5594081. eCollection 2021. Biomed Res Int. 2021. PMID: 33860037 Free PMC article.
-
Comprehensively Identifying the Key tRNA-Derived Fragments and Investigating Their Function in Gastric Cancer Processes.Onco Targets Ther. 2020 Oct 28;13:10931-10943. doi: 10.2147/OTT.S266130. eCollection 2020. Onco Targets Ther. 2020. PMID: 33149609 Free PMC article.
-
Identifying Differentially Expressed tRNA-Derived Small Fragments as a Biomarker for the Progression and Metastasis of Colorectal Cancer.Dis Markers. 2022 Jan 6;2022:2646173. doi: 10.1155/2022/2646173. eCollection 2022. Dis Markers. 2022. PMID: 35035608 Free PMC article.
-
Recent Insights Into Wnt-Related tRNA-Derived Fragments (tRFs) in Human Diseases.J Cell Biochem. 2025 Jan;126(1):e30702. doi: 10.1002/jcb.30702. J Cell Biochem. 2025. PMID: 39835731 Review.
-
tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections.Theranostics. 2021 Jan 1;11(1):461-469. doi: 10.7150/thno.51963. eCollection 2021. Theranostics. 2021. PMID: 33391486 Free PMC article. Review.
Cited by
-
tRNA-derived fragments (tRFs) in cancer.J Cell Commun Signal. 2023 Mar;17(1):47-54. doi: 10.1007/s12079-022-00690-2. Epub 2022 Aug 29. J Cell Commun Signal. 2023. PMID: 36036848 Free PMC article. Review.
-
tRNA-derived RNA fragments in cancer: current status and future perspectives.J Hematol Oncol. 2020 Sep 4;13(1):121. doi: 10.1186/s13045-020-00955-6. J Hematol Oncol. 2020. PMID: 32887641 Free PMC article. Review.
-
A novel 3'tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer.Cell Death Dis. 2022 May 18;13(5):471. doi: 10.1038/s41419-022-04930-6. Cell Death Dis. 2022. PMID: 35585048 Free PMC article.
-
Role of tRNA-derived small RNAs(tsRNAs) in the diagnosis and treatment of malignant tumours.Cell Commun Signal. 2023 Jul 21;21(1):178. doi: 10.1186/s12964-023-01199-w. Cell Commun Signal. 2023. PMID: 37480078 Free PMC article. Review.
-
The Emerging Function and Promise of tRNA-Derived Small RNAs in Cancer.J Cancer. 2024 Jan 27;15(6):1642-1656. doi: 10.7150/jca.89219. eCollection 2024. J Cancer. 2024. PMID: 38370372 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

