Inhibited Lipophagy Suppresses Lipid Metabolism in Zebrafish Liver Cells
- PMID: 31496957
- PMCID: PMC6713122
- DOI: 10.3389/fphys.2019.01077
Inhibited Lipophagy Suppresses Lipid Metabolism in Zebrafish Liver Cells
Abstract
Lipophagy degrades lipid droplets (LDs) through the lysosomal degradative pathway, thus plays important roles in regulating lipid metabolism in mammals. However, information on the existence and functions of lipophagy in fish lipid metabolism is still limited. In the present study, we confirmed the existence of lipophagy by observing the structures of LDs sequestered in autophagic vacuoles in the zebrafish liver cell line (ZFL) via electronic microscopy. Moreover, starved cells increased the mRNA expression of the microtubule-associated protein 1A/1B light chain 3 beta (LC3), which is a marker protein for autophagy and protein conversion from LC3-I to LC3-II. Inhibiting autophagy with chloroquine increased significantly the LDs content and decreased fatty acid β-oxidation and esterification activities in the ZFL cells cultured in the fed state. Furthermore, inhibiting autophagy function downregulated the mRNA expression of the genes and their proteins related to lipid metabolism. Altogether, the present study verified the existence of lipophagy and its essential regulatory roles in lipid metabolism in fish cells.
Keywords: esterification; fatty acid β-oxidation; lipid metabolism; lipophagy; zebrafish liver cells.
Figures
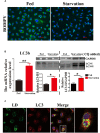
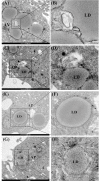


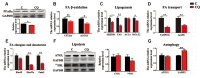
Similar articles
-
Lipophagy-derived fatty acids undergo extracellular efflux via lysosomal exocytosis.Autophagy. 2021 Mar;17(3):690-705. doi: 10.1080/15548627.2020.1728097. Epub 2020 Feb 19. Autophagy. 2021. PMID: 32070194 Free PMC article.
-
AmAtg2B-Mediated Lipophagy Regulates Lipolysis of Pupae in Apis mellifera.Int J Mol Sci. 2023 Jan 20;24(3):2096. doi: 10.3390/ijms24032096. Int J Mol Sci. 2023. PMID: 36768418 Free PMC article.
-
[Notch1 inhibits the mechanistic role of STING signaling to regulate hepatocyte lipophagy in nonalcoholic steatohepatitis].Zhonghua Gan Zang Bing Za Zhi. 2023 Aug 20;31(8):827-834. doi: 10.3760/cma.j.cn501113-20230208-00042. Zhonghua Gan Zang Bing Za Zhi. 2023. PMID: 37723064 Chinese.
-
Lipophagy and liver disease: New perspectives to better understanding and therapy.Biomed Pharmacother. 2018 Jan;97:339-348. doi: 10.1016/j.biopha.2017.07.168. Epub 2017 Nov 6. Biomed Pharmacother. 2018. PMID: 29091883 Review.
-
The ménage à trois of autophagy, lipid droplets and liver disease.Autophagy. 2022 Jan;18(1):50-72. doi: 10.1080/15548627.2021.1895658. Epub 2021 Apr 2. Autophagy. 2022. PMID: 33794741 Free PMC article. Review.
Cited by
-
Retraction of: PHTF2 regulates lipids metabolism in gastric cancer.Aging (Albany NY). 2024 Apr 30;16(8):7507. doi: 10.18632/aging.205842. Epub 2024 Apr 30. Aging (Albany NY). 2024. PMID: 38688693 Free PMC article. No abstract available.
-
Modulation of Autophagy: A Novel "Rejuvenation" Strategy for the Aging Liver.Oxid Med Cell Longev. 2021 Feb 10;2021:6611126. doi: 10.1155/2021/6611126. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 33628363 Free PMC article. Review.
-
Selective autophagy: the rise of the zebrafish model.Autophagy. 2021 Nov;17(11):3297-3305. doi: 10.1080/15548627.2020.1853382. Epub 2020 Dec 15. Autophagy. 2021. PMID: 33228439 Free PMC article. Review.
-
Current Understanding on the Role of Lipids in Macrophages and Associated Diseases.Int J Mol Sci. 2022 Dec 29;24(1):589. doi: 10.3390/ijms24010589. Int J Mol Sci. 2022. PMID: 36614031 Free PMC article. Review.
-
Microlipophagy from Simple to Complex Eukaryotes.Cells. 2025 Jan 18;14(2):141. doi: 10.3390/cells14020141. Cells. 2025. PMID: 39851569 Free PMC article. Review.
References
-
- Du H., Heur M., Duanmu M., Grabowski G. A., Hui D. Y., Witte D. P., et al. (2001). Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J. Lipid Res. 42 489–500. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

