Evolution of Yin and Yang isoforms of a chromatin remodeling subunit precedes the creation of two genes
- PMID: 31498079
- PMCID: PMC6752949
- DOI: 10.7554/eLife.48119
Evolution of Yin and Yang isoforms of a chromatin remodeling subunit precedes the creation of two genes
Abstract
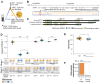
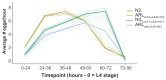
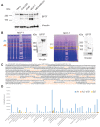
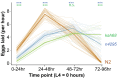
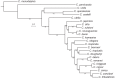
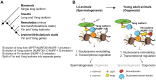
Genes can encode multiple isoforms, broadening their functions and providing a molecular substrate to evolve phenotypic diversity. Evolution of isoform function is a potential route to adapt to new environments. Here we show that de novo, beneficial alleles in the nurf-1 gene became fixed in two laboratory lineages of C. elegans after isolation from the wild in 1951, before methods of cryopreservation were developed. nurf-1 encodes an ortholog of BPTF, a large (>300 kD) multidomain subunit of the NURF chromatin remodeling complex. Using CRISPR-Cas9 genome editing and transgenic rescue, we demonstrate that in C. elegans, nurf-1 has split into two, largely non-overlapping isoforms (NURF-1.D and NURF-1.B, which we call Yin and Yang, respectively) that share only two of 26 exons. Both isoforms are essential for normal gametogenesis but have opposite effects on male/female gamete differentiation. Reproduction in hermaphrodites, which involves production of both sperm and oocytes, requires a balance of these opposing Yin and Yang isoforms. Transgenic rescue and genetic position of the fixed mutations suggest that different isoforms are modified in each laboratory strain. In a related clade of Caenorhabditis nematodes, the shared exons have duplicated, resulting in the split of the Yin and Yang isoforms into separate genes, each containing approximately 200 amino acids of duplicated sequence that has undergone accelerated protein evolution following the duplication. Associated with this duplication event is the loss of two additional nurf-1 transcripts, including the long-form transcript and a newly identified, highly expressed transcript encoded by the duplicated exons. We propose these lost transcripts are non-functional side products necessary to transcribe the Yin and Yang transcripts in the same cells. Our work demonstrates how gene sharing, through the production of multiple isoforms, can precede the creation of new, independent genes.
Keywords: C. elegans; Isoform evolution; adaptive conflict; alternative transcripts; chromatin remodeling; evolutionary biology; other Caenorhabditis species.
© 2019, Xu et al.
Conflict of interest statement
WX, LL, YZ, LS, IF, JM, RE, PM No competing interests declared
Figures















References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

