High-throughput sequencing analysis of microbial community diversity in response to indica and japonica bar-transgenic rice paddy soils
- PMID: 31498816
- PMCID: PMC6733487
- DOI: 10.1371/journal.pone.0222191
High-throughput sequencing analysis of microbial community diversity in response to indica and japonica bar-transgenic rice paddy soils
Abstract
Potential environmental risks of genetically modified (GM) crops have raised concerns. To better understand the effect of transgenic rice on the bacterial community in paddy soil, a field experiment was carried out using pairs of rice varieties from two subspecies (indica and japonica) containing bar transgene with herbicide resistance and their parental conventional rice. The 16S rRNA gene of soil genomic DNA from different soil layers at the maturity stage was sequenced using high-throughput sequencing on the Illumina MiSeq platform to explore the microbial community diversity among different rice soils. There were no significant differences in diversity indices between transgenic japonica rice and its sister conventional rice (japonica pair) among different soil layers, but, significant differences was observed between transgenic indica rice and its conventional rice (indica pair) in the topsoil layer around concentrated rice roots according to the ace diversity index. Though the japonica rice soil and indica rice soil were shared several key genera, including Rivibacter, Anaeromyxobacter, Roseomonas, Geobacter, Thiobacillus, Clostridium, and Desulfobulbus, the primary bacterial genera in indica rice soil were different from those in japonica rice. Synechococcus and Dechloromonas were present in japonica rice samples, while Chloronema, Flexibacter, and Blastocatella were observed in indica rice soil. Moreover, the abundance of genera between GM and non-GM varieties in japonica rice was significantly different from indica rice, and several bacterial communities influenced these differences. Anaerovorax was more abundant in transgenic japonica rice soil than conventional rice soil, while it was deficient in transgenic indica rice soil compared to conventional rice soil, and opposite responses to Deferrisoma were in that of indica rice. Thus, we concluded that transgenic indica and japonica rice had different effects on soil bacteria compared with their corresponding sister conventional rice. However, these composition and abundance difference only occurred for a few genera but had no effect on the primary genera and soil characteristics were mainly contributed to these differences. Thus, differences in bacterial community structure can be ignored when evaluating the impacts of transgenic rice in the complex soil microenvironment.
Conflict of interest statement
The authors have declared that no competing interests exist.
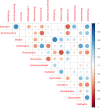
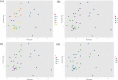
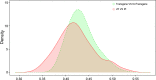
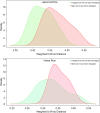
Figures



References
-
- James C. Global Status of Commercialized Biotech/GM Crops in 2017:Biotech Crop Adoption Surges as Economic Benefits Accumulate in 22 Years: ISAAA; 2018 [cited 2018 Sep 22]. Available from: http://www.isaaa.org/resources/publications/annualreport/2017/default.asp.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

