Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate
- PMID: 31500178
- PMCID: PMC6770254
- DOI: 10.3390/ijms20184394
Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate
Abstract
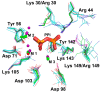
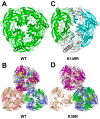
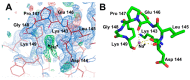
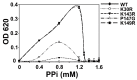
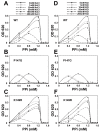
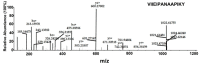
All living things have pyrophosphatases that hydrolyze pyrophosphate and release energy. This energetically favorable reaction drives many energetically unfavorable reactions. An accepted catalytic model of pyrophosphatase shows that a water molecule activated by two divalent cations (M1 and M2) within the catalytic center can attack pyrophosphate in an SN2 mechanism and thus hydrolyze the molecule. However, our co-crystal structure of Acinetobacter baumannii pyrophosphatase with pyrophosphate shows that a water molecule from the solvent may, in fact, be the actual catalytic water. In the co-crystal structure of the wild-type pyrophosphatase with pyrophosphate, the electron density of the catalytic centers of each monomer are different from one another. This indicates that pyrophosphates in the catalytic center are dynamic. Our mass spectroscopy results have identified a highly conserved lysine residue (Lys30) in the catalytic center that is phosphorylated, indicating that the enzyme could form a phosphoryl enzyme intermediate during hydrolysis. Mutation of Lys30 to Arg abolished the activity of the enzyme. In the structure of the apo wild type enzyme, we observed that a Na+ ion is coordinated by residues within a loop proximal to the catalytic center. Therefore, we mutated three key residues within the loop (K143R, P147G, and K149R) and determined Na+ and K+-induced inhibition on their activities. Compared to the wild type enzyme, P147G is most sensitive to these cations, whereas K143R was inactive and K149R showed no change in activity. These data indicate that monovalent cations could play a role in down-regulating pyrophosphatase activity in vivo. Overall, our results reveal new aspects of pyrophosphatase catalysis and could assist in the design of specific inhibitors of Acinetobacter baumannii growth.
Keywords: Acinetobacter baumannii; catalytic mechanism; crystal structure; inorganic pyrophosphatase; lysine phosphorylation; mass spectrometry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A new method to investigate the catalytic mechanism of YhdE pyrophosphatase by using a pyrophosphate fluorescence probe.Sci Rep. 2017 Aug 15;7(1):8169. doi: 10.1038/s41598-017-08368-1. Sci Rep. 2017. PMID: 28811554 Free PMC article.
-
Structure/function analysis of a dUTPase: catalytic mechanism of a potential chemotherapeutic target.J Mol Biol. 1999 Apr 30;288(2):275-87. doi: 10.1006/jmbi.1999.2680. J Mol Biol. 1999. PMID: 10329142
-
Role of the potassium/lysine cationic center in catalysis and functional asymmetry in membrane-bound pyrophosphatases.Biochem J. 2018 Mar 26;475(6):1141-1158. doi: 10.1042/BCJ20180071. Biochem J. 2018. PMID: 29519958
-
Role of water in processes of energy transduction: Ca2+-transport ATPase and inorganic pyrophosphatase.Biochem Soc Symp. 1985;50:97-125. Biochem Soc Symp. 1985. PMID: 2428374 Review.
-
Inorganic pyrophosphatases: one substrate, three mechanisms.FEBS Lett. 2013 Jun 27;587(13):1863-9. doi: 10.1016/j.febslet.2013.05.003. Epub 2013 May 16. FEBS Lett. 2013. PMID: 23684653 Review.
Cited by
-
Characterization of a family I inorganic pyrophosphatase from Legionella pneumophila Philadelphia 1.Acta Crystallogr F Struct Biol Commun. 2023 Oct 1;79(Pt 10):257-266. doi: 10.1107/S2053230X23008002. Epub 2023 Sep 20. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 37728609 Free PMC article.
References
-
- Nelson D., Cox M. Lehninger Principles of Biochemistry. 3rd ed. Worth Publishers; New York, NY, USA: 2000. p. 937.
-
- Reginald H., Charles M. Grisham Biochemistry. 4th ed. Cengage Learning, Brooks/Cole; Belmont, CA, USA: 2010.
-
- Van Wazer J.R., Griffith E.J., Mccullough J.F. Structure and Properties of the Condensed Phosphates. VII. Hydrolytic Degradation of Pyro- and Tripolyphosphate. J. Am. Chem. Soc. 1955;77:287–291. doi: 10.1021/ja01607a011. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

