Origin and diversification of Xanthomonas citri subsp. citri pathotypes revealed by inclusive phylogenomic, dating, and biogeographic analyses
- PMID: 31500575
- PMCID: PMC6734499
- DOI: 10.1186/s12864-019-6007-4
Origin and diversification of Xanthomonas citri subsp. citri pathotypes revealed by inclusive phylogenomic, dating, and biogeographic analyses
Abstract
Background: Xanthomonas citri subsp. citri pathotypes cause bacterial citrus canker, being responsible for severe agricultural losses worldwide. The A pathotype has a broad host spectrum, while A* and Aw are more restricted both in hosts and in geography. Two previous phylogenomic studies led to contrasting well-supported clades for sequenced genomes of these pathotypes. No extensive biogeographical or divergence dating analytic approaches have been so far applied to available genomes.
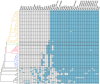
Results: Based on a larger sampling of genomes than in previous studies (including six new genomes sequenced by our group, adding to a total of 95 genomes), phylogenomic analyses resulted in different resolutions, though overall indicating that A + AW is the most likely true clade. Our results suggest the high degree of recombination at some branches and the fast diversification of lineages are probable causes for this phylogenetic blurring effect. One of the genomes analyzed, X. campestris pv. durantae, was shown to be an A* strain; this strain has been reported to infect a plant of the family Verbenaceae, though there are no reports of any X. citri subsp. citri pathotypes infecting any plant outside the Citrus genus. Host reconstruction indicated the pathotype ancestor likely had plant hosts in the family Fabaceae, implying an ancient jump to the current Rutaceae hosts. Extensive dating analyses indicated that the origin of X. citri subsp. citri occurred more recently than the main phylogenetic splits of Citrus plants, suggesting dispersion rather than host-directed vicariance as the main driver of geographic expansion. An analysis of 120 pathogenic-related genes revealed pathotype-associated patterns of presence/absence.
Conclusions: Our results provide novel insights into the evolutionary history of X. citri subsp. citri as well as a sound phylogenetic foundation for future evolutionary and genomic studies of its pathotypes.
Keywords: Biogeography; Divergence dating; Genome evolution; Phylogenomics; Recombination.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Civerolo EL. Bacterial canker disease of citrus [Xanthomonas campestris] Journal of the Rio Grande Valley Horticultural Society. 1984;37:127–145.
-
- Lee HA. Further data on the susceptibility of rutaceous plants to citrus-canker. J Agric Res. 1918;15:661–665.
-
- Bitancourt AA. O Cancro Cítrico. Biológico. 1957;23:101–111.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

