Evolution of a 72-Kilobase Cointegrant, Conjugative Multiresistance Plasmid in Community-Associated Methicillin-Resistant Staphylococcus aureus Isolates from the Early 1990s
- PMID: 31501140
- PMCID: PMC6811413
- DOI: 10.1128/AAC.01560-19
Evolution of a 72-Kilobase Cointegrant, Conjugative Multiresistance Plasmid in Community-Associated Methicillin-Resistant Staphylococcus aureus Isolates from the Early 1990s
Abstract
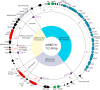
Horizontal transfer of plasmids encoding antimicrobial resistance and virulence determinants has been instrumental in Staphylococcus aureus evolution, including the emergence of community-associated methicillin-resistant S. aureus (CA-MRSA). In the early 1990s, the first CA-MRSA strain isolated in Western Australia (WA), WA-5, encoded cadmium, tetracycline, and penicillin resistance genes on plasmid pWBG753 (∼30 kb). WA-5 and pWBG753 appeared only briefly in WA; however, fusidic acid resistance plasmids related to pWBG753 were also present in the first European CA-MRSA isolates at the time. Here, we characterize a 72-kb conjugative plasmid, pWBG731, present in multiresistant WA-5-like clones from the same period. pWBG731 was a cointegrant formed from pWBG753 and a pWBG749 family conjugative plasmid. pWBG731 carried mupirocin, trimethoprim, cadmium, and penicillin resistance genes. The stepwise evolution of pWBG731 likely occurred through the combined actions of IS257, IS257-dependent miniature inverted-repeat transposable elements (MITEs), and the BinL resolution system of the β-lactamase transposon Tn552 An evolutionarily intermediate ∼42-kb nonconjugative plasmid, pWBG715, possessed the same resistance genes as pWBG731 but retained an integrated copy of the small tetracycline resistance plasmid pT181. IS257 likely facilitated the replacement of pT181 with conjugation genes on pWBG731, thus enabling autonomous transfer. Like conjugative plasmid pWBG749, pWBG731 also mobilized nonconjugative plasmids carrying oriT mimics. It seems likely that pWBG731 represents the product of multiple recombination events between the WA-5 pWBG753 plasmid and other mobile genetic elements present in indigenous community-associated methicillin-sensitive S. aureus (CA-MSSA) isolates. The molecular evolution of pWBG731 saliently illustrates how diverse mobile genetic elements can together facilitate rapid accrual and horizontal dissemination of multiresistance in S. aureus CA-MRSA.
Keywords: CA-MRSA; IS257; MITEs; Staphylococcus aureus; Tn552; conjugation; mupirocin; plasmid; resolvase; trimethoprim.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Genetic analysis of high-level mupirocin resistance in the ST80 clone of community-associated meticillin-resistant Staphylococcus aureus.J Med Microbiol. 2010 Feb;59(Pt 2):193-199. doi: 10.1099/jmm.0.013268-0. Epub 2009 Oct 15. J Med Microbiol. 2010. PMID: 19833783
-
Multidrug-resistant clones of community-associated meticillin-resistant Staphylococcus aureus isolated from Chinese children and the resistance genes to clindamycin and mupirocin.J Med Microbiol. 2012 Sep;61(Pt 9):1240-1247. doi: 10.1099/jmm.0.042663-0. Epub 2012 May 17. J Med Microbiol. 2012. PMID: 22595913
-
Emergence of high-level mupirocin resistance in methicillin-resistant Staphylococcus aureus in Western Australia.J Hosp Infect. 1994 Mar;26(3):157-65. doi: 10.1016/0195-6701(94)90038-8. J Hosp Infect. 1994. PMID: 7911481
-
Multiresistant Staphylococcus aureus: genetics and evolution of epidemic Australian strains.J Antimicrob Chemother. 1988 Apr;21 Suppl C:19-39. doi: 10.1093/jac/21.suppl_c.19. J Antimicrob Chemother. 1988. PMID: 2838448 Review.
-
The evolution of Staphylococcus aureus.Infect Genet Evol. 2008 Dec;8(6):747-63. doi: 10.1016/j.meegid.2008.07.007. Epub 2008 Jul 29. Infect Genet Evol. 2008. PMID: 18718557 Review.
Cited by
-
Recombination in Bacterial Genomes: Evolutionary Trends.Toxins (Basel). 2023 Sep 12;15(9):568. doi: 10.3390/toxins15090568. Toxins (Basel). 2023. PMID: 37755994 Free PMC article. Review.
-
Efflux Pump Mediated Antimicrobial Resistance by Staphylococci in Health-Related Environments: Challenges and the Quest for Inhibition.Antibiotics (Basel). 2021 Dec 7;10(12):1502. doi: 10.3390/antibiotics10121502. Antibiotics (Basel). 2021. PMID: 34943714 Free PMC article. Review.
-
The Plasmidomic Landscape of Clinical Methicillin-Resistant Staphylococcus aureus Isolates from Malaysia.Antibiotics (Basel). 2023 Apr 9;12(4):733. doi: 10.3390/antibiotics12040733. Antibiotics (Basel). 2023. PMID: 37107095 Free PMC article.
-
Functional Roles and Genomic Impact of Miniature Inverted-Repeat Transposable Elements (MITEs) in Prokaryotes.Genes (Basel). 2024 Mar 3;15(3):328. doi: 10.3390/genes15030328. Genes (Basel). 2024. PMID: 38540387 Free PMC article. Review.
-
Prevalence and genomic insights into type III-A CRISPR-Cas system acquisition in global Staphylococcus argenteus strains.Front Cell Infect Microbiol. 2025 Jul 28;15:1644286. doi: 10.3389/fcimb.2025.1644286. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40792102 Free PMC article.
References
-
- Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis 42:647–656. doi:10.1086/499815. - DOI - PubMed
-
- Boan P, Tan HL, Pearson J, Coombs G, Heath CH, Robinson JO. 2015. Epidemiological, clinical, outcome and antibiotic susceptibility differences between PVL positive and PVL negative Staphylococcus aureus infections in Western Australia: a case control study. BMC Infect Dis 15:10. doi:10.1186/s12879-014-0742-6. - DOI - PMC - PubMed
-
- Coombs GW, Daly DA, Pearson JC, Nimmo GR, Collignon PJ, McLaws ML, Robinson JO, Turnidge JD, Australian Group on Antimicrobial Resistance. 2014. Community-onset Staphylococcus aureus Surveillance Programme annual report, 2012. Commun Dis Intell Q Rep 38:E59–E69. - PubMed
-
- Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O’Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591. doi:10.1534/g3.111.000760. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

