Development of Transposon Mutagenesis for Chlamydia muridarum
- PMID: 31501283
- PMCID: PMC6832062
- DOI: 10.1128/JB.00366-19
Development of Transposon Mutagenesis for Chlamydia muridarum
Abstract
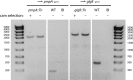
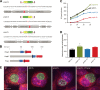
Functional genetic analysis of Chlamydia has been a challenge due to the historical genetic intractability of Chlamydia, although recent advances in chlamydial genetic manipulation have begun to remove these barriers. Here, we report the development of the Himar C9 transposon system for Chlamydia muridarum, a mouse-adapted Chlamydia species that is widely used in Chlamydia infection models. We demonstrate the generation and characterization of an initial library of 33 chloramphenicol (Cam)-resistant, green fluorescent protein (GFP)-expressing C. muridarum transposon mutants. The majority of the mutants contained single transposon insertions spread throughout the C. muridarum chromosome. In all, the library contained 31 transposon insertions in coding open reading frames (ORFs) and 7 insertions in intergenic regions. Whole-genome sequencing analysis of 17 mutant clones confirmed the chromosomal locations of the insertions. Four mutants with transposon insertions in glgB, pmpI, pmpA, and pmpD were investigated further for in vitro and in vivo phenotypes, including growth, inclusion morphology, and attachment to host cells. The glgB mutant was shown to be incapable of complete glycogen biosynthesis and accumulation in the lumen of mutant inclusions. Of the 3 pmp mutants, pmpI was shown to have the most pronounced growth attenuation defect. This initial library demonstrates the utility and efficacy of stable, isogenic transposon mutants for C. muridarum The generation of a complete library of C. muridarum mutants will ultimately enable comprehensive identification of the functional genetic requirements for Chlamydia infection in vivoIMPORTANCE Historical issues with genetic manipulation of Chlamydia have prevented rigorous functional genetic characterization of the ∼1,000 genes in chlamydial genomes. Here, we report the development of a transposon mutagenesis system for C. muridarum, a mouse-adapted Chlamydia species that is widely used for in vivo investigations of chlamydial pathogenesis. This advance builds on the pioneering development of this system for C. trachomatis We demonstrate the generation of an initial library of 33 mutants containing stable single or double transposon insertions. Using these mutant clones, we characterized in vitro phenotypes associated with genetic disruptions in glycogen biosynthesis and three polymorphic outer membrane proteins.
Keywords: Chlamydia muridarum; glycogen; pmp; transposon mutagenesis.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Johnson NB, Hayes LD, Brown K, Hoo EC, Ethier KA, Centers for Disease Control and Prevention. 2014. CDC National Health Report: leading causes of morbidity and mortality and associated behavioral risk and protective factors—United States, 2005–2013. MMWR Suppl 63:3–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

