Yeast PAF1 complex counters the pol III accumulation and replication stress on the tRNA genes
- PMID: 31501524
- PMCID: PMC6733944
- DOI: 10.1038/s41598-019-49316-5
Yeast PAF1 complex counters the pol III accumulation and replication stress on the tRNA genes
Abstract
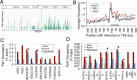
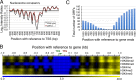
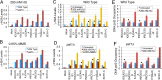
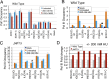
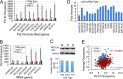
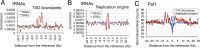
The RNA polymerase (pol) III transcribes mostly short, house-keeping genes, which produce stable, non-coding RNAs. The tRNAs genes, highly transcribed by pol III in vivo are known replication fork barriers. One of the transcription factors, the PAF1C (RNA polymerase II associated factor 1 complex) is reported to associate with pol I and pol II and influence their transcription. We found low level PAF1C occupancy on the yeast pol III-transcribed genes, which is not correlated with nucleosome positions, pol III occupancy and transcription. PAF1C interacts with the pol III transcription complex and causes pol III loss from the genes under replication stress. Genotoxin exposure causes pol III but not Paf1 loss from the genes. In comparison, Paf1 deletion leads to increased occupancy of pol III, γ-H2A and DNA pol2 in gene-specific manner. Paf1 restricts the accumulation of pol III by influencing the pol III pause on the genes, which reduces the pol III barrier to the replication fork progression.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II.Mol Cell Biol. 2006 Apr;26(8):3135-48. doi: 10.1128/MCB.26.8.3135-3148.2006. Mol Cell Biol. 2006. PMID: 16581788 Free PMC article.
-
Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress.Genes Dev. 2016 Feb 1;30(3):337-54. doi: 10.1101/gad.273813.115. Epub 2016 Jan 21. Genes Dev. 2016. PMID: 26798134 Free PMC article.
-
Interactome of the yeast RNA polymerase III transcription machinery constitutes several chromatin modifiers and regulators of the genes transcribed by RNA polymerase II.Gene. 2019 Jun 20;702:205-214. doi: 10.1016/j.gene.2018.12.037. Epub 2018 Dec 26. Gene. 2019. PMID: 30593915
-
Maf1, a general negative regulator of RNA polymerase III in yeast.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):376-84. doi: 10.1016/j.bbagrm.2012.11.004. Epub 2012 Nov 28. Biochim Biophys Acta. 2013. PMID: 23201230 Review.
-
Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):320-329. doi: 10.1016/j.bbagrm.2018.01.011. Epub 2018 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29378333 Review.
Cited by
-
Characterization of Tau95 led to the identification of a four-subunit TFIIIC complex in trypanosomatid parasites.Appl Microbiol Biotechnol. 2024 Dec;108(1):109. doi: 10.1007/s00253-023-12903-8. Epub 2024 Jan 10. Appl Microbiol Biotechnol. 2024. PMID: 38204130 Free PMC article.
-
Participation of TFIIIB Subunit Brf1 in Transcription Regulation in the Human Pathogen Leishmania major.Genes (Basel). 2021 Feb 16;12(2):280. doi: 10.3390/genes12020280. Genes (Basel). 2021. PMID: 33669344 Free PMC article.
-
Transcription-dependent enrichment of the yeast FACT complex influences nucleosome dynamics on the RNA polymerase III-transcribed genes.RNA. 2020 Dec 4;27(3):273-90. doi: 10.1261/rna.077974.120. Online ahead of print. RNA. 2020. PMID: 33277439 Free PMC article.
-
High-resolution Sequencing Reveals that the Paf1 Complex May be a Conserved Transcription Elongation Factor for Eukaryotic RNA Polymerase I.J Mol Biol. 2025 Sep 1;437(17):169220. doi: 10.1016/j.jmb.2025.169220. Epub 2025 May 19. J Mol Biol. 2025. PMID: 40398673
-
A regulatory phosphorylation site on Mec1 controls chromatin occupancy of RNA polymerases during replication stress.EMBO J. 2021 Nov 2;40(21):e108439. doi: 10.15252/embj.2021108439. Epub 2021 Sep 27. EMBO J. 2021. PMID: 34569643 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

