Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome
- PMID: 31501537
- PMCID: PMC6879825
- DOI: 10.1038/s41564-019-0550-2
Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome
Abstract
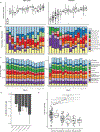
Hospitalized preterm infants receive frequent and often prolonged exposures to antibiotics because they are vulnerable to infection. It is not known whether the short-term effects of antibiotics on the preterm infant gut microbiota and resistome persist after discharge from neonatal intensive care units. Here, we use complementary metagenomic, culture-based and machine learning techniques to study the gut microbiota and resistome of antibiotic-exposed preterm infants during and after hospitalization, and we compare these readouts to antibiotic-naive healthy infants sampled synchronously. We find a persistently enriched gastrointestinal antibiotic resistome, prolonged carriage of multidrug-resistant Enterobacteriaceae and distinct antibiotic-driven patterns of microbiota and resistome assembly in extremely preterm infants that received early-life antibiotics. The collateral damage of early-life antibiotic treatment and hospitalization in preterm infants is long lasting. We urge the development of strategies to reduce these consequences in highly vulnerable neonatal populations.
Figures



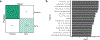
Comment in
-
Early-life antibiotic use and gut microbiota.Nat Rev Genet. 2019 Nov;20(11):630. doi: 10.1038/s41576-019-0174-7. Nat Rev Genet. 2019. PMID: 31520076 No abstract available.
References
Methods References
-
- Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

