scds: computational annotation of doublets in single-cell RNA sequencing data
- PMID: 31501871
- PMCID: PMC7703774
- DOI: 10.1093/bioinformatics/btz698
scds: computational annotation of doublets in single-cell RNA sequencing data
Abstract
Motivation: Single-cell RNA sequencing (scRNA-seq) technologies enable the study of transcriptional heterogeneity at the resolution of individual cells and have an increasing impact on biomedical research. However, it is known that these methods sometimes wrongly consider two or more cells as single cells, and that a number of so-called doublets is present in the output of such experiments. Treating doublets as single cells in downstream analyses can severely bias a study's conclusions, and therefore computational strategies for the identification of doublets are needed.
Results: With scds, we propose two new approaches for in silico doublet identification: Co-expression based doublet scoring (cxds) and binary classification based doublet scoring (bcds). The co-expression based approach, cxds, utilizes binarized (absence/presence) gene expression data and, employing a binomial model for the co-expression of pairs of genes, yields interpretable doublet annotations. bcds, on the other hand, uses a binary classification approach to discriminate artificial doublets from original data. We apply our methods and existing computational doublet identification approaches to four datasets with experimental doublet annotations and find that our methods perform at least as well as the state of the art, at comparably little computational cost. We observe appreciable differences between methods and across datasets and that no approach dominates all others. In summary, scds presents a scalable, competitive approach that allows for doublet annotation of datasets with thousands of cells in a matter of seconds.
Availability and implementation: scds is implemented as a Bioconductor R package (doi: 10.18129/B9.bioc.scds).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures

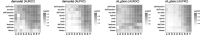
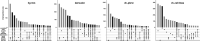

References
-
- Chen T., Guestrin C. (2016) XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’16. ACM, NY, USA, pp. 785–794.

