Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution
- PMID: 31502753
- PMCID: PMC6832101
- DOI: 10.15252/embr.201847016
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution
Abstract
Most mosquito-borne flaviviruses, including Zika virus (ZIKV), Dengue virus (DENV), and West Nile virus (WNV), produce long non-coding subgenomic RNAs (sfRNAs) in infected cells that link to pathogenicity and immune evasion. Until now, the structural characterization of these lncRNAs remains limited. Here, we studied the 3D structures of individual and combined subdomains of sfRNAs, and visualized the accessible 3D conformational spaces of complete sfRNAs from DENV2, ZIKV, and WNV by small angle X-ray scattering (SAXS) and computational modeling. The individual xrRNA1s and xrRNA2s adopt similar structures in solution as the crystal structure of ZIKV xrRNA1, and all xrRNA1-2s form compact structures with reduced flexibility. While the DB12 of DENV2 is extended, the DB12s of ZIKV and WNV are compact due to the formation of intertwined double pseudoknots. All 3' stem-loops (3'SLs) share similar rod-like structures. Complete sfRNAs are extended and sample a large conformational space in solution. Our work not only provides structural insight into the function of flavivirus sfRNAs, but also highlights strategies of visualizing other lncRNAs in solution by SAXS and computational methods.
Keywords: 3D structure; computational modeling; long non-coding RNA; small angle X-ray scattering; subgenomic flavivirus RNA.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
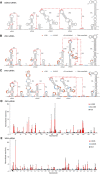
- A–C
Secondary structure of the complete sfRNAs of DENV2 (A), ZIKV (B), and WNV (C). The stem‐loop structures (SLI and SLII), SL3 of WNV, pseudo‐dumbbell (ψ‐DB1) structure of ZIKV, duplicated dumbbell structures (DB1, DB2), and the essential 3′ SL are indicated. Sequences involved in pseudoknots (PK1, PK2, PK3, PK4) formation are indicated with red lines.
- D, E
SHAPE analysis of the complete sfRNAs of ZIKV (D) and WNV (E). Nucleotides with SHAPE reactivity greater than 0.85 are labeled in red, and moderately reactive site (0.4–0.85) is labeled in cyan. Unlabeled sites have low or no SHAPE reactivity (< 0.4). The SHAPE reactivity was also annotated onto the corresponding secondary structures in (B) and (C). Nucleotides which SHAPE reactivity was not determined are labeled in gray in (B) and (C).
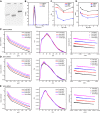
- A
Tris‐borate‐magnesium (TBM) polyacrylamide gel for sfRNAs of DENV2, ZIKV, and WNV, respectively.
- B
The distribution of the light scattering by sfRNAs as a function of particle radius in solution.
- C–E
The scattering curves (left), PDDFs (middle), and guinier plots (right) for sfRNAs of DENV2 (C), ZIKV (D), and WNV (E) at various Mg2+ concentrations.
- F, G
R g (F) and D max (G) of sfRNAs derived from guinier plots and PDDF calculation were plotted as a function of Mg2+ concentration.
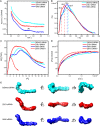
- A–D
The scattering profiles (A), pair distance distribution functions (PDDFs) (B), dimensionless Kratky plots (C), and Porod‐Debye plots (D) of the complete sfRNAs of DENV2 (cyan line), ZIKV (red line), and WNV (blue line). The fine features of P1 in the scattering profiles in (A) arise from helical interstrand pair distance correlation. The PDDF profiles in (B) were calculated using GNOM (q max = 0.3). The dimensionless Kratky plot and Porod‐Debye plot for ZIKV xrRNA1 (red dashed line) were included in (C) and (D) for comparison, indicating that the structures of sfRNAs are more extended and open than that of ZIKV xrRNA1.
- E
The ab initio shape envelopes of the complete sfRNAs shown in three views. The spatial resolution of the envelopes is 21 Å.

- A–C
For each RNA, forty independent ab initio shape reconstructions were generated with DAMMIN in slow mode and superimposed with DAMSEL and DAMSUP with NSD values as indicated. Four of 40 individual reconstructions are shown for each RNA in two different orientations, suggesting significant divergence among the shape reconstructions. All 40 superimposed models in each case were averaged and filtered with DAMAVER and DAMFILT to generate the final shape envelopes shown in the boxes on the left.
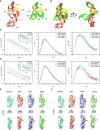
- A
Crystal structures of ZIKV xrRNA1 (left, PDB code: 5TPY) and MVEV xrRNA2 (right, PDB code: 4PQV).
- B
Two views of the overlay of ZIKV xrRNA1 and MVEV xrRNA2 crystal structures.
- C, D
The scattering profiles (left), the PDDFs (middle), and the dimensionless Kratky plots (right) of individual xrRNA1s (C) and xrRNA2s (D) of DENV2 (cyan), ZIKV (red), WNV (blue), and MVEV (green). The insets in (C) and (D) show the Guinier regions of the respective scattering profiles with a linear fit line. The back‐calculated scattering profile of ZIKV‐xrRNA1 crystal structure (black, open circle) can be nicely fitted onto its experimental SAXS data (red line) in (C), while the back‐calculated scattering profile of MVEV‐xrRNA2 crystal structure (black, open circle) fits poorly to its experimental SAXS data (green line) in (D). The asymmetric PDDFs of xrRNA1 (C) and xrRNA2 (D) indicate slightly elongated molecules with asymmetric shapes. The dimensionless Kratky plots of xrRNA1 (C) and xrRNA2 (D) of DENV2, ZIKV, WNV, and MVEV suggest all individual xrRNA1s and xrRNA2s are well‐folded and of reduced flexibility in solution.
- E, F
The ab initio shape envelopes of the individual xrRNA1s (E) and xrRNA2s (F) of DENV2 (cyan), ZIKV (red), WNV (blue), and MVEV (green) are fitted with the homologous atomic models built by ModeRNA.

Sequence alignment of xrRNA1s and xrRNA2s from DENV2, ZIKV, WNV, and MVEV. The consensus sequences are highlighted in yellow. Sequences in the respective helical or loop regions are indicated at the top.
Representative fits of the theoretical scattering curves of DAMMIN bead models and ModeRNA atomic models to the experimental SAXS data of xrRNA1s (top row) and xrRNA2s (bottom row) of DENV2, ZIKV, WNV, and MVEV, respectively.
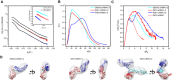
- A–C
The scattering profiles (A), the PDDFs (B), and the dimensionless Kratky plots (C) of xrRNA1‐2s from DENV2 (cyan), ZIKV (red), and WNV (blue). The inset in (A) shows the guinier regions of the respective scattering profiles with linear fit lines. The dimensionless Kratky plot of ZIKV xrRNA1 (red dashed line) was included in (C) for comparison, indicating that the structures of xrRNA1‐2s are all well‐folded.
- D
The ab initio shape envelopes of the xrRNA1‐2s of DENV2 (left), ZIKV (middle), and WNV (right). The atomic models from rigid‐body modeling by Xplor‐NIH are superimposed onto the respective envelopes. The back‐calculated scattering profiles of the atomic models were fitted to the respective experimental scattering profiles in (A).
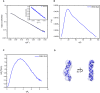
- A–C
Scattering profile (A), PDDFs (B), and dimensionless Kratky plots (C) of SL3 of WNV. The inset in (A) is the guinier region with fitting line of the scattering profile.
- D
The de novo atomic model by Rosetta was superimposed onto the shape envelope of WNV‐SL3. The back‐calculated scattering profile of the atomic model (black line) was fitted to the experimental SAXS data of WNV‐SL3 (blue line).

- A–C
The scattering profiles (A), the PDDFs (B), and the dimensionless Kratky plots (C) of DB12s from DENV2 (cyan), ZIKV (red), and WNV (blue). The inset in (A) shows the Guinier regions of the respective scattering profiles with linear fit lines. The dimensionless Kratky plot of ZIKV xrRNA1 (red dashed line) was included in (C) for comparison, indicating that the structures of DB12s of ZIKV and WNV are well‐folded, but that of DB12 of DENV2 is partially folded. The Kratky plots for the two PK3 mutants of DB12s, the ZIKV‐DB12‐PK3Δ (green) and WNV DB12‐PK3Δ (black), respectively, are also included for comparison, suggesting reduced compactness upon PK3 mutations.
- D
The ab initio shape envelopes of the DB12s of DENV2 (left), ZIKV (middle), and WNV (right) in two views. The atomic models from de novo structure modeling by Rosetta were superimposed onto the respective envelopes. The back‐calculated scattering profiles of the de novo atomic models are fitted to the respective experimental scattering profiles in (A).
- E
The ab initio shape envelopes of the PK3 mutants of ZIKV DB12s, the ZIKV‐DB12‐PK3Δ (left) and WNV DB12‐PK3Δ (right) in two views, suggesting open conformations upon PK3 mutations.
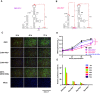
- A, B
Mutational schemes of the two PK3 mutants, ZIKV‐PK3^ (A) and ZIKV‐PK3* (B), mapping to the secondary structures of the DB12 region.
- C
E protein expression of ZIKV and the mutants showed by IFA.
- D
The progeny virus RNA in cultured supernatants of ZIKV and the mutants was detected by qRT–PCR in three technical replicates. Two‐way ANOVA and Dunnett's multiple comparison test were used for statistical analysis. The error bars represent the standard deviation. ***P < 0.001, ****P < 0.0001.
- E
The luciferase activity of the ZIKV‐Rep and the mutants was measured at different time points after transfection in three technical replicates. The error bars represent the standard deviation.

- A–C
The scattering profiles (A), the PDDFs (B), and the dimensionless Kratky plots (C) of 3′SLs from DENV2 (cyan), ZIKV (red), WNV (blue), and WNV‐3′SLM (438UAG441A→438GUC441U) mutant (green). The inset in (A) shows the guinier regions of the respective scattering profiles with linear fit lines.
- D
The ab initio shape envelopes of the 3′SLs of DENV2 (left), ZIKV (middle), and WNV (right), WNV‐3′SLM (green). The atomic models from de novo structure modeling by Rosetta except for 3′SLM of WNV are superimposed onto the respective envelopes. The back‐calculated scattering profiles of the de novo atomic models are fitted to the respective experimental scattering profiles in (A).
- E
Illustration of the three binding sites (orange asterisks) of WNV‐3′SL to eEF1A (pink ellipse), the potential pseudoknot is also indicated with red dash line.
- F
The structure of ribosome‐bound eEF1A (PDB: 5LZS) is compared with the envelope and atomic model of WNV‐3′SL side by side.
- G
Sequence alignment of DENV2‐, ZIKV‐ and WNV‐3′SL. The sequences involved in the potential pseudoknot between the sHP and long SL of WNV 3′SL were indicated with # sign at the top.
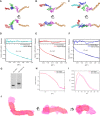
- A–F
Using the available atomic models of the individual subdomains of each sfRNA, all‐atom atomic models can be constructed and refined by rigid‐body‐modeling against SAXS data using Xplor‐NIH. The back‐calculated scattering curves of the best fit models for DENV2 (A), ZIKV (B), and WNV (C) were fitted to the experimental scattering curves of DENV2 (D), ZIKV (E), and WNV (F), respectively.
- G
Native PAGE gel for ZIKV sfRNA and ZIKV sfRNA‐LNA complex, the latter shows obvious shift retardation relative to ZIKV sfRNA in PAGE gel, indicating that the LNA binding affects overall structure of ZIKV sfRNA.
- H, I
The differences in scattering profiles (H) and pair distance distribution functions (PDDFs) (I) between ZIKV sfRNA‐LNA complex (magenta) and ZIKV sfRNAs (red) indicate that LNA binding affects the overall structure of ZIKV sfRNA significantly. The PDDF profiles in (I) were calculated using GNOM (q max = 0.3).
- J
Three views of the overlay of the shape envelopes of ZIKV sfRNA‐LNA (magenta) onto that of ZIKV sfRNA (red).
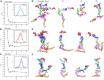
- A–C
The fitting chi‐square is plotted against the ensemble size for complete sfRNAs of DENV2 (A), ZIKV (B), and WNV (C), suggesting optimized minimal structural ensembles with 3, 3,3 conformers, respectively. The respective insets show the distribution of the minimal ensembles and the initial pools of 10,000 random conformers as a function of Rg.
- D–F
The models of the selected minimal structural ensembles (N e = 3) that can reproduce the scattering curves of the complete sfRNAs of DENV2 (D), ZIKV (E), and WNV (F) are shown. In each panel, the left are the three individual models in each ensemble overlaid on top of the DB1 domains, the right are the three individual models in each ensemble displayed separately. The xrRNA1, xrRNA2, SL3 of WNV, ψ‐DB1 and DB1, DB2, and 3′SL domains are colored in red, blue, cyan, green, magenta, and orange, respectively.
References
-
- Nugent EK, Nugent AK, Nugent R, Nugent K (2017) Zika virus: epidemiology, pathogenesis and human disease. Am J Med Sci 353: 466–473 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 2016YFA0500700/The National Key Research and Development Program of China/International
- 2016YFD0500304/The National Key Research and Development Program of China/International
- 2017ZX09101005/The National Key Research and Development Program of China/International
- 2018ZX09711003/The National Key Research and Development Program of China/International
- 81522025/National Natural Science Foundation of China/International
- 81621005/National Natural Science Foundation of China/International
- 31770190/National Natural Science Foundation of China/International
- China Youth 1000-Talent Program of the State Council of China, Tsinghua University Initiative Scientific Research Program/International
- Beijing Advanced Innovation Center for Structural Biology/International
- Tsinghua-Peking Joint Center for Life Sciences/International
LinkOut - more resources
Full Text Sources

