Antisense regulation of atrial natriuretic peptide expression
- PMID: 31503546
- PMCID: PMC6795498
- DOI: 10.1172/jci.insight.130978
Antisense regulation of atrial natriuretic peptide expression
Abstract
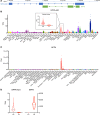
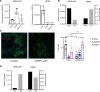
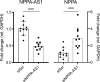
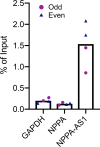
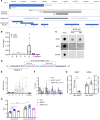
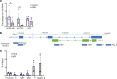
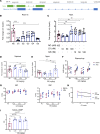
The cardiac hormone atrial natriuretic peptide (ANP) is a central regulator of blood volume and a therapeutic target in hypertension and heart failure. Enhanced ANP activity in such conditions through inhibition of the degradative enzyme neprilysin has shown clinical efficacy but is complicated by consequences of simultaneous accumulation of a heterogeneous array of other hormones. Targets for specific ANP enhancement have not been available. Here, we describe a cis-acting antisense transcript (NPPA-AS1), which negatively regulates ANP expression in human cardiomyocytes. We show that NPPA-AS1 regulates ANP expression via facilitating NPPA repressor RE1-silencing transcription factor (REST) binding to its promoter, rather than forming an RNA duplex with ANP mRNA. Expression of ANP mRNA and NPPA-AS1 was increased and correlated in isolated strained human cardiomyocytes and in hearts from patients with advanced heart failure. Further, inhibition of NPPA-AS1 in vitro and in vivo resulted in increased myocardial expression of ANP, increased circulating ANP, increased renal cGMP, and lower blood pressure. The effects of NPPA-AS1 inhibition on NPPA expression in human cardiomyocytes were further marked under cell-strain conditions. Collectively, these results implicate the antisense transcript NPPA-AS1 as part of a physiologic self-regulatory ANP circuit and a viable target for specific ANP augmentation.
Keywords: Cardiology; Heart failure; Noncoding RNAs; Transcription.
Conflict of interest statement
Figures

References
-
- Sergeeva IA, Christoffels VM. Regulation of expression of atrial and brain natriuretic peptide, biomarkers for heart development and disease. Biochim Biophys Acta. 2013;1832(12):2403–2413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

