The phage gene wmk is a candidate for male killing by a bacterial endosymbiont
- PMID: 31504075
- PMCID: PMC6736233
- DOI: 10.1371/journal.ppat.1007936
The phage gene wmk is a candidate for male killing by a bacterial endosymbiont
Abstract
Wolbachia are the most widespread maternally-transmitted bacteria in the animal kingdom. Their global spread in arthropods and varied impacts on animal physiology, evolution, and vector control are in part due to parasitic drive systems that enhance the fitness of infected females, the transmitting sex of Wolbachia. Male killing is one common drive mechanism wherein the sons of infected females are selectively killed. Despite decades of research, the gene(s) underlying Wolbachia-induced male killing remain unknown. Here using comparative genomic, transgenic, and cytological approaches in fruit flies, we identify a candidate gene in the eukaryotic association module of Wolbachia prophage WO, termed WO-mediated killing (wmk), which transgenically causes male-specific lethality during early embryogenesis and cytological defects typical of the pathology of male killing. The discovery of wmk establishes new hypotheses for the potential role of phage genes in sex-specific lethality, including the control of arthropod pests and vectors.
Conflict of interest statement
J.I.P. and Seth R.B. are listed as inventors on a patent related to potential applications of wmk in arthropods.
Figures

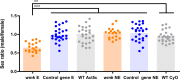
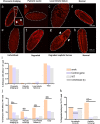

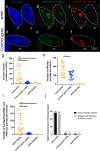

References
Publication types
MeSH terms
Substances
Grants and funding
- WT094664MA/WT_/Wellcome Trust/United Kingdom
- R21 HD086833/HD/NICHD NIH HHS/United States
- R01 AI139154/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- S10 OD021630/OD/NIH HHS/United States
- R21 AI133522/AI/NIAID NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- R01 AI132581/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U24 DK059637/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- R00 GM114714/GM/NIGMS NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- 281668/ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

