Analysis of several key factors influencing deep learning-based inter-residue contact prediction
- PMID: 31504181
- PMCID: PMC7703788
- DOI: 10.1093/bioinformatics/btz679
Analysis of several key factors influencing deep learning-based inter-residue contact prediction
Abstract
Motivation: Deep learning has become the dominant technology for protein contact prediction. However, the factors that affect the performance of deep learning in contact prediction have not been systematically investigated.
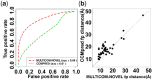
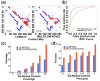
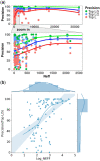
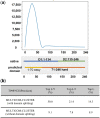
Results: We analyzed the results of our three deep learning-based contact prediction methods (MULTICOM-CLUSTER, MULTICOM-CONSTRUCT and MULTICOM-NOVEL) in the CASP13 experiment and identified several key factors [i.e. deep learning technique, multiple sequence alignment (MSA), distance distribution prediction and domain-based contact integration] that influenced the contact prediction accuracy. We compared our convolutional neural network (CNN)-based contact prediction methods with three coevolution-based methods on 75 CASP13 targets consisting of 108 domains. We demonstrated that the CNN-based multi-distance approach was able to leverage global coevolutionary coupling patterns comprised of multiple correlated contacts for more accurate contact prediction than the local coevolution-based methods, leading to a substantial increase of precision by 19.2 percentage points. We also tested different alignment methods and domain-based contact prediction with the deep learning contact predictors. The comparison of the three methods showed deeper sequence alignments and the integration of domain-based contact prediction with the full-length contact prediction improved the performance of contact prediction. Moreover, we demonstrated that the domain-based contact prediction based on a novel ab initio approach of parsing domains from MSAs alone without using known protein structures was a simple, fast approach to improve contact prediction. Finally, we showed that predicting the distribution of inter-residue distances in multiple distance intervals could capture more structural information and improve binary contact prediction.
Availability and implementation: https://github.com/multicom-toolbox/DNCON2/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures

Similar articles
-
Improving deep learning-based protein distance prediction in CASP14.Bioinformatics. 2021 Oct 11;37(19):3190-3196. doi: 10.1093/bioinformatics/btab355. Bioinformatics. 2021. PMID: 33961009 Free PMC article.
-
Improving protein tertiary structure prediction by deep learning and distance prediction in CASP14.Proteins. 2022 Jan;90(1):58-72. doi: 10.1002/prot.26186. Epub 2021 Jul 27. Proteins. 2022. PMID: 34291486 Free PMC article.
-
Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13.Proteins. 2019 Dec;87(12):1165-1178. doi: 10.1002/prot.25697. Epub 2019 Apr 25. Proteins. 2019. PMID: 30985027 Free PMC article.
-
Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction.Int J Mol Sci. 2021 Jun 2;22(11):6032. doi: 10.3390/ijms22116032. Int J Mol Sci. 2021. PMID: 34199677 Free PMC article. Review.
-
Exploring novel ANGICon-EIPs through ameliorated peptidomics techniques: Can deep learning strategies as a core breakthrough in peptide structure and function prediction?Food Res Int. 2023 Dec;174(Pt 1):113640. doi: 10.1016/j.foodres.2023.113640. Epub 2023 Oct 27. Food Res Int. 2023. PMID: 37986483 Review.
Cited by
-
How much metagenome data is needed for protein structure prediction: The advantages of targeted approach from the ecological and evolutionary perspectives.Imeta. 2022 Mar 6;1(1):e9. doi: 10.1002/imt2.9. eCollection 2022 Mar. Imeta. 2022. PMID: 38867727 Free PMC article. Review.
-
Improving Protein Secondary Structure Prediction by Deep Language Models and Transformer Networks.Methods Mol Biol. 2025;2867:43-53. doi: 10.1007/978-1-0716-4196-5_3. Methods Mol Biol. 2025. PMID: 39576574
-
Deep learning methods in protein structure prediction.Comput Struct Biotechnol J. 2020 Jan 22;18:1301-1310. doi: 10.1016/j.csbj.2019.12.011. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32612753 Free PMC article. Review.
-
Tertiary structure assessment at CASP15.Proteins. 2023 Dec;91(12):1616-1635. doi: 10.1002/prot.26593. Epub 2023 Sep 25. Proteins. 2023. PMID: 37746927 Free PMC article.
-
Accurate structure prediction of biomolecular interactions with AlphaFold 3.Nature. 2024 Jun;630(8016):493-500. doi: 10.1038/s41586-024-07487-w. Epub 2024 May 8. Nature. 2024. PMID: 38718835 Free PMC article.
References
-
- Altschuh D. et al. (1988) Coordinated amino acid changes in homologous protein families. Protein Eng., 2, 193–199. - PubMed
-
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54 (Pt 5), 905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

