A Rare Mutation in SPLUNC1 Affects Bacterial Adherence and Invasion in Meningococcal Disease
- PMID: 31504285
- PMCID: PMC7201419
- DOI: 10.1093/cid/ciz600
A Rare Mutation in SPLUNC1 Affects Bacterial Adherence and Invasion in Meningococcal Disease
Erratum in
-
Correction to: A Rare Mutation in SPLUNC1 Affects Bacterial Adherence and Invasion in Meningococcal Disease.Clin Infect Dis. 2022 Aug 24;75(1):184. doi: 10.1093/cid/ciac263. Clin Infect Dis. 2022. PMID: 35608985 Free PMC article. No abstract available.
Abstract
Background: Neisseria meningitidis (Nm) is a nasopharyngeal commensal carried by healthy individuals. However, invasive infections occurs in a minority of individuals, with devastating consequences. There is evidence that common polymorphisms are associated with invasive meningococcal disease (IMD), but the contributions of rare variants other than those in the complement system have not been determined.
Methods: We identified familial cases of IMD in the UK meningococcal disease study and the European Union Life-Threatening Infectious Disease Study. Candidate genetic variants were identified by whole-exome sequencing of 2 patients with familial IMD. Candidate variants were further validated by in vitro assays.
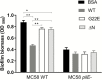
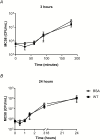
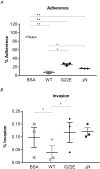
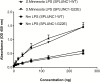
Results: Exomes of 2 siblings with IMD identified a novel heterozygous missense mutation in BPIFA1/SPLUNC1. Sequencing of 186 other nonfamilial cases identified another unrelated IMD patient with the same mutation. SPLUNC1 is an innate immune defense protein expressed in the nasopharyngeal epithelia; however, its role in invasive infections is unknown. In vitro assays demonstrated that recombinant SPLUNC1 protein inhibits biofilm formation by Nm, and impedes Nm adhesion and invasion of human airway cells. The dominant negative mutant recombinant SPLUNC1 (p.G22E) showed reduced antibiofilm activity, increased meningococcal adhesion, and increased invasion of cells, compared with wild-type SPLUNC1.
Conclusions: A mutation in SPLUNC1 affecting mucosal attachment, biofilm formation, and invasion of mucosal epithelial cells is a new genetic cause of meningococcal disease.
Keywords: human genetics; meningococcal disease; mucosal immunity; sepsis; severe infectious disease.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures


References
-
- Yazdankhah SP, Caugant DA. Neisseria meningitidis: an overview of the carriage state. J Med Microbiol 2004; 53:821–32. - PubMed
-
- Virji M. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat Rev Microbiol 2009; 7:274–86. - PubMed
-
- Mace SE. Acute bacterial meningitis. Emerg Med Clin North Am 2008; 26:281–317, viii. - PubMed
-
- Pace D, Pollard AJ. Meningococcal disease: clinical presentation and sequelae. Vaccine 2012; 30(Suppl 2):B3–9. - PubMed

