The Genomic Substrate for Adaptive Radiation: Copy Number Variation across 12 Tribes of African Cichlid Species
- PMID: 31504491
- PMCID: PMC6795240
- DOI: 10.1093/gbe/evz185
The Genomic Substrate for Adaptive Radiation: Copy Number Variation across 12 Tribes of African Cichlid Species
Abstract
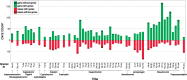
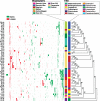
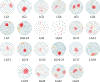
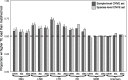
The initial sequencing of five cichlid genomes revealed an accumulation of genetic variation, including extensive copy number variation in cichlid lineages particularly those that have undergone dramatic evolutionary radiation. Gene duplication has the potential to generate substantial molecular substrate for the origin of evolutionary novelty. We use array-based comparative heterologous genomic hybridization to identify copy number variation events (CNVEs) for 168 samples representing 53 cichlid species including the 5 species for which full genome sequence is available. We identify an average of 50-100 CNVEs per individual. For those species represented by multiple samples, we identify 150-200 total CNVEs suggesting a substantial amount of intraspecific variation. For these species, only ∼10% of the detected CNVEs are fixed. Hierarchical clustering of species according to CNVE data recapitulates phylogenetic relationships fairly well at both the tribe and radiation level. Although CNVEs are detected on all linkage groups, they tend to cluster in "hotspots" and are likely to contain and be flanked by transposable elements. Furthermore, we show that CNVEs impact functional categories of genes with potential roles in adaptive phenotypes that could reasonably promote divergence and speciation in the cichlid clade. These data contribute to a more complete understanding of the molecular basis for adaptive natural selection, speciation, and evolutionary radiation.
Keywords: adaptive radiation; cichlid; copy number variation; gene duplication; genomic architecture.
Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution 2019. This work is written by US Government employees and is in the public domain in the US.
Figures

Similar articles
-
The genomic substrate for adaptive radiation in African cichlid fish.Nature. 2014 Sep 18;513(7518):375-381. doi: 10.1038/nature13726. Epub 2014 Sep 3. Nature. 2014. PMID: 25186727 Free PMC article.
-
Gene duplication in an African cichlid adaptive radiation.BMC Genomics. 2014 Feb 26;15:161. doi: 10.1186/1471-2164-15-161. BMC Genomics. 2014. PMID: 24571567 Free PMC article.
-
African cichlid fish: a model system in adaptive radiation research.Proc Biol Sci. 2006 Aug 22;273(1597):1987-98. doi: 10.1098/rspb.2006.3539. Proc Biol Sci. 2006. PMID: 16846905 Free PMC article. Review.
-
Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation.Mol Ecol. 2013 Feb;22(3):787-98. doi: 10.1111/mec.12023. Epub 2012 Oct 12. Mol Ecol. 2013. PMID: 23057853
-
Genetic Variation and Hybridization in Evolutionary Radiations of Cichlid Fishes.Annu Rev Anim Biosci. 2021 Feb 16;9:55-79. doi: 10.1146/annurev-animal-061220-023129. Epub 2020 Nov 16. Annu Rev Anim Biosci. 2021. PMID: 33197206 Review.
Cited by
-
Identification of High-Confidence Structural Variants in Domesticated Rainbow Trout Using Whole-Genome Sequencing.Front Genet. 2021 Feb 25;12:639355. doi: 10.3389/fgene.2021.639355. eCollection 2021. Front Genet. 2021. PMID: 33732289 Free PMC article.
-
Epigenetic Regulation and Environmental Sex Determination in Cichlid Fishes.Sex Dev. 2021;15(1-3):93-107. doi: 10.1159/000517197. Epub 2021 Aug 25. Sex Dev. 2021. PMID: 34433170 Free PMC article. Review.
-
The comparative genomic landscape of adaptive radiation in crater lake cichlid fishes.Mol Ecol. 2021 Feb;30(4):955-972. doi: 10.1111/mec.15774. Epub 2021 Jan 6. Mol Ecol. 2021. PMID: 33305470 Free PMC article.
-
Extensive genome-wide duplications in the eastern oyster (Crassostrea virginica).Philos Trans R Soc Lond B Biol Sci. 2021 May 24;376(1825):20200164. doi: 10.1098/rstb.2020.0164. Epub 2021 Apr 5. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33813893 Free PMC article.
-
Correspondence of aCGH and long-read genome assembly for detection of copy number differences: A proof-of-concept with cichlid genomes.PLoS One. 2021 Oct 7;16(10):e0258193. doi: 10.1371/journal.pone.0258193. eCollection 2021. PLoS One. 2021. PMID: 34618847 Free PMC article.
References
-
- Altschul SF, et al. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

