Validity of polygenic risk scores: are we measuring what we think we are?
- PMID: 31504522
- PMCID: PMC7013150
- DOI: 10.1093/hmg/ddz205
Validity of polygenic risk scores: are we measuring what we think we are?
Abstract
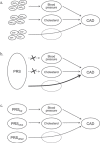
Polygenic risk scores (PRSs) have become the standard for quantifying genetic liability in the prediction of disease risks. PRSs are generally constructed as weighted sum scores of risk alleles using effect sizes from genome-wide association studies as their weights. The construction of PRSs is being improved with more appropriate selection of independent single-nucleotide polymorphisms (SNPs) and optimized estimation of their weights but is rarely reflected upon from a theoretical perspective, focusing on the validity of the risk score. Borrowing from psychometrics, this paper discusses the validity of PRSs and introduces the three main types of validity that are considered in the evaluation of tests and measurements: construct, content, and criterion validity. This introduction is followed by a discussion of three topics that challenge the validity of PRS, namely, their claimed independence of clinical risk factors, the consequences of relaxing SNP inclusion thresholds and the selection of SNP weights. This discussion of the validity of PRS reminds us that we need to keep questioning if weighted sums of risk alleles are measuring what we think they are in the various scenarios in which PRSs are used and that we need to keep exploring alternative modeling strategies that might better reflect the underlying biological pathways.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Torkamani A., Wineinger N.E. and Topol E.J. (2018) The personal and clinical utility of polygenic risk scores. Nat Rev Genet., 19, 581–590. - PubMed
-
- Nelson R.M., Pettersson M.E. and Carlborg O. (2013) A century after Fisher: time for a new paradigm in quantitative genetics. Trend Genet., 29, 669–676. - PubMed
-
- Seddon J.M., George S., Rosner B. and Klein M.L. (2006) CFH gene variant, Y402H, and smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered., 61, 157–165. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

