IRES-mediated cap-independent translation, a path leading to hidden proteome
- PMID: 31504667
- PMCID: PMC6884710
- DOI: 10.1093/jmcb/mjz091
IRES-mediated cap-independent translation, a path leading to hidden proteome
Abstract
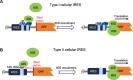
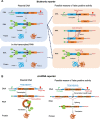
Most eukaryotic mRNAs are translated in a cap-dependent fashion; however, under stress conditions, the cap-independent translation driven by internal ribosomal entry sites (IRESs) can serve as an alternative mechanism for protein production. Many IRESs have been discovered from viral or cellular mRNAs to promote ribosome assembly and initiate translation by recruiting different trans-acting factors. Although the mechanisms of translation initiation driven by viral IRESs are relatively well understood, the existence of cellular IRESs is still under debate due to the limitations of translation reporter systems used to assay IRES activities. A recent screen identified > 1000 putative IRESs from viral and human mRNAs, expanding the scope and mechanism for cap-independent translation. Additionally, a large number of circular RNAs lacking free ends were identified in eukaryotic cells, many of which are found to be translated through IRESs. These findings suggest that IRESs may play a previously unappreciated role in driving translation of the new type of mRNA, implying a hidden proteome produced from cap-independent translation.
Keywords: IRES; ITAF; bicistronic system; circular RNA; translation.
© The Author(s) (2019). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures

Similar articles
-
IRESbase: A Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites.Genomics Proteomics Bioinformatics. 2020 Apr;18(2):129-139. doi: 10.1016/j.gpb.2020.03.001. Epub 2020 Jun 6. Genomics Proteomics Bioinformatics. 2020. PMID: 32512182 Free PMC article.
-
Alternative ways to think about cellular internal ribosome entry.J Biol Chem. 2010 Sep 17;285(38):29033-8. doi: 10.1074/jbc.R110.150532. Epub 2010 Jun 24. J Biol Chem. 2010. PMID: 20576611 Free PMC article. Review.
-
A circular split nanoluciferase reporter for validating and screening putative internal ribosomal entry site elements.RNA. 2024 Oct 16;30(11):1529-1540. doi: 10.1261/rna.080008.124. RNA. 2024. PMID: 39103230
-
Sequence features of viral and human Internal Ribosome Entry Sites predictive of their activity.PLoS Comput Biol. 2017 Sep 18;13(9):e1005734. doi: 10.1371/journal.pcbi.1005734. eCollection 2017 Sep. PLoS Comput Biol. 2017. PMID: 28922394 Free PMC article.
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
Cited by
-
SFPQ promotes RAS-mutant cancer cell growth by modulating 5'-UTR mediated translational control of CK1α.NAR Cancer. 2022 Sep 27;4(3):zcac027. doi: 10.1093/narcan/zcac027. eCollection 2022 Sep. NAR Cancer. 2022. PMID: 36177382 Free PMC article.
-
The Critical Roles of Circular RNAs in Basic Research and Clinical Application of Female Reproductive-Related Diseases.Reprod Sci. 2023 May;30(5):1421-1434. doi: 10.1007/s43032-022-01070-2. Epub 2022 Oct 5. Reprod Sci. 2023. PMID: 36197632 Review.
-
The dark proteome: translation from noncanonical open reading frames.Trends Cell Biol. 2022 Mar;32(3):243-258. doi: 10.1016/j.tcb.2021.10.010. Epub 2021 Nov 26. Trends Cell Biol. 2022. PMID: 34844857 Free PMC article. Review.
-
Structure and function of type IV IRES in picornaviruses: a systematic review.Front Microbiol. 2024 May 24;15:1415698. doi: 10.3389/fmicb.2024.1415698. eCollection 2024. Front Microbiol. 2024. PMID: 38855772 Free PMC article.
-
Newly discovered mechanisms that mediate tumorigenesis and tumour progression: circRNA-encoded proteins.J Cell Mol Med. 2023 Jun;27(12):1609-1620. doi: 10.1111/jcmm.17751. Epub 2023 Apr 18. J Cell Mol Med. 2023. PMID: 37070530 Free PMC article. Review.
References
-
- Baerenfaller K., Grossmann J., Grobei M.A., et al. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

