Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection
- PMID: 31504706
- PMCID: PMC6812734
- DOI: 10.1093/jxb/erz332
Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection
Abstract
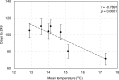
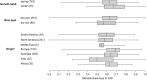
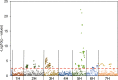
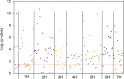
Single-marker genome-wide association studies (GWAS) have successfully detected associations between single nucleotide polymorphisms (SNPs) and agronomic traits such as flowering time and grain yield in barley. However, the analysis of individual SNPs can only account for a small proportion of genetic variation, and can only provide limited knowledge on gene network interactions. Gene-based GWAS approaches provide enormous opportunity both to combine genetic information and to examine interactions among genetic variants. Here, we revisited a previously published phenotypic and genotypic data set of 895 barley varieties grown in two years at four different field locations in Australia. We employed statistical models to examine gene-phenotype associations, as well as two-way epistasis analyses to increase the capability to find novel genes that have significant roles in controlling flowering time in barley. Genetic associations were tested between flowering time and corresponding genotypes of 174 putative flowering time-related genes. Gene-phenotype association analysis detected 113 genes associated with flowering time in barley, demonstrating the unprecedented power of gene-based analysis. Subsequent two-way epistasis analysis revealed 19 pairs of gene×gene interactions involved in controlling flowering time. Our study demonstrates that gene-based association approaches can provide higher capacity for future crop improvement to increase crop performance and adaptation to different environments.
Keywords: Barley; GWAS; epistasis; flowering time; gene-set association analysis; heritability; next-generation sequencing; phenology; target capture; target enrichment.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures
Similar articles
-
Modelling the genetic architecture of flowering time control in barley through nested association mapping.BMC Genomics. 2015 Apr 12;16(1):290. doi: 10.1186/s12864-015-1459-7. BMC Genomics. 2015. PMID: 25887319 Free PMC article.
-
Hybridisation-based target enrichment of phenology genes to dissect the genetic basis of yield and adaptation in barley.Plant Biotechnol J. 2019 May;17(5):932-944. doi: 10.1111/pbi.13029. Epub 2018 Dec 6. Plant Biotechnol J. 2019. PMID: 30407713 Free PMC article.
-
Genome-wide association studies for agronomical traits in a world wide spring barley collection.BMC Plant Biol. 2012 Jan 27;12:16. doi: 10.1186/1471-2229-12-16. BMC Plant Biol. 2012. PMID: 22284310 Free PMC article.
-
Major flowering time genes of barley: allelic diversity, effects, and comparison with wheat.Theor Appl Genet. 2021 Jul;134(7):1867-1897. doi: 10.1007/s00122-021-03824-z. Epub 2021 May 9. Theor Appl Genet. 2021. PMID: 33969431 Free PMC article. Review.
-
Detecting epistasis in human complex traits.Nat Rev Genet. 2014 Nov;15(11):722-33. doi: 10.1038/nrg3747. Epub 2014 Sep 9. Nat Rev Genet. 2014. PMID: 25200660 Review.
Cited by
-
Assisted Gene Flow Management to Climate Change in the Annual Legume Lupinus angustifolius L.: From Phenotype to Genotype.Evol Appl. 2025 Mar 6;18(3):e70087. doi: 10.1111/eva.70087. eCollection 2025 Mar. Evol Appl. 2025. PMID: 40059886 Free PMC article.
-
Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan.Plants (Basel). 2023 Jun 19;12(12):2375. doi: 10.3390/plants12122375. Plants (Basel). 2023. PMID: 37376001 Free PMC article.
-
Haplotype-Based, Genome-Wide Association Study Reveals Stable Genomic Regions for Grain Yield in CIMMYT Spring Bread Wheat.Front Genet. 2020 Dec 3;11:589490. doi: 10.3389/fgene.2020.589490. eCollection 2020. Front Genet. 2020. PMID: 33335539 Free PMC article.
-
Genome architecture and diverged selection shaping pattern of genomic differentiation in wild barley.Plant Biotechnol J. 2023 Jan;21(1):46-62. doi: 10.1111/pbi.13917. Epub 2022 Nov 16. Plant Biotechnol J. 2023. PMID: 36054248 Free PMC article.
-
Population origin determines the adaptive potential for the advancement of flowering onset in Lupinus angustifolius L. (Fabaceae).Evol Appl. 2022 Nov 29;16(1):62-73. doi: 10.1111/eva.13510. eCollection 2023 Jan. Evol Appl. 2022. PMID: 36699122 Free PMC article.
References
-
- Baum M, von Korff M, Guo P, Lakew B, Udupa SM, Sayed H, Choumane W, Grando S, Ceccarelli S. 2007. Molecular approaches and breeding strategies for drought tolerance in barley. In: Varshney RK, Tuberosa R, eds. Genomic assisted crop improvement, vol. 2, genomics applications in crops. Dordrecht, The Netherlands: Springer, 51–79.
-
- Blázquez MA, Weigel D. 2000. Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. - PubMed
-
- Blümel M, Dally N, Jung C. 2015. Flowering time regulation in crops—what did we learn from Arabidopsis? Current Opinion in Biotechnology 32, 121–129. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

