The Evolution of Reverse Gyrase Suggests a Nonhyperthermophilic Last Universal Common Ancestor
- PMID: 31504731
- PMCID: PMC6878951
- DOI: 10.1093/molbev/msz180
The Evolution of Reverse Gyrase Suggests a Nonhyperthermophilic Last Universal Common Ancestor
Abstract
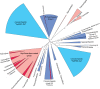
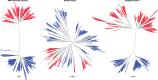
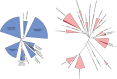
Reverse gyrase (RG) is the only protein found ubiquitously in hyperthermophilic organisms, but absent from mesophiles. As such, its simple presence or absence allows us to deduce information about the optimal growth temperature of long-extinct organisms, even as far as the last universal common ancestor of extant life (LUCA). The growth environment and gene content of the LUCA has long been a source of debate in which RG often features. In an attempt to settle this debate, we carried out an exhaustive search for RG proteins, generating the largest RG data set to date. Comprising 376 sequences, our data set allows for phylogenetic reconstructions of RG with unprecedented size and detail. These RG phylogenies are strikingly different from those of universal proteins inferred to be present in the LUCA, even when using the same set of species. Unlike such proteins, RG does not form monophyletic archaeal and bacterial clades, suggesting RG emergence after the formation of these domains, and/or significant horizontal gene transfer. Additionally, the branch lengths separating archaeal and bacterial groups are very short, inconsistent with the tempo of evolution from the time of the LUCA. Despite this, phylogenies limited to archaeal RG resolve most archaeal phyla, suggesting predominantly vertical evolution since the time of the last archaeal ancestor. In contrast, bacterial RG indicates emergence after the last bacterial ancestor followed by significant horizontal transfer. Taken together, these results suggest a nonhyperthermophilic LUCA and bacterial ancestor, with hyperthermophily emerging early in the evolution of the archaeal and bacterial domains.
Keywords: LUCA; evolution; hyperthermophiles; phylogeny; reverse gyrase.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
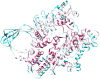
Similar articles
-
Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers.Archaea. 2007 May;2(2):83-93. doi: 10.1155/2006/582916. Archaea. 2007. PMID: 17350929 Free PMC article.
-
DNA supercoiling and temperature adaptation: A clue to early diversification of life?J Mol Evol. 1999 Oct;49(4):439-52. doi: 10.1007/pl00006567. J Mol Evol. 1999. PMID: 10486002
-
Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes.BMC Evol Biol. 2003 Jan 6;3:2. doi: 10.1186/1471-2148-3-2. Epub 2003 Jan 6. BMC Evol Biol. 2003. PMID: 12515582 Free PMC article.
-
The Last Universal Common Ancestor of Ribosome-Encoding Organisms: Portrait of LUCA.J Mol Evol. 2024 Oct;92(5):550-583. doi: 10.1007/s00239-024-10186-9. Epub 2024 Aug 19. J Mol Evol. 2024. PMID: 39158619 Review.
-
Origin and evolution of DNA topoisomerases.Biochimie. 2007 Apr;89(4):427-46. doi: 10.1016/j.biochi.2006.12.009. Epub 2007 Jan 4. Biochimie. 2007. PMID: 17293019 Review.
Cited by
-
The methanogen core and pangenome: conservation and variability across biology's growth temperature extremes.DNA Res. 2023 Feb 1;30(1):dsac048. doi: 10.1093/dnares/dsac048. DNA Res. 2023. PMID: 36454681 Free PMC article.
-
Adaptive laboratory evolution of a thermophile toward a reduced growth temperature optimum.Front Microbiol. 2023 Oct 12;14:1265216. doi: 10.3389/fmicb.2023.1265216. eCollection 2023. Front Microbiol. 2023. PMID: 37901835 Free PMC article.
-
A Place for Viruses on the Tree of Life.Front Microbiol. 2021 Jan 14;11:604048. doi: 10.3389/fmicb.2020.604048. eCollection 2020. Front Microbiol. 2021. PMID: 33519747 Free PMC article. Review.
-
Carl Woese: Still ahead of our time.mLife. 2022 Dec 14;1(4):359-367. doi: 10.1002/mlf2.12049. eCollection 2022 Dec. mLife. 2022. PMID: 38818481 Free PMC article. No abstract available.
-
Proteome-wide 3D structure prediction provides insights into the ancestral metabolism of ancient archaea and bacteria.Nat Commun. 2022 Dec 21;13(1):7861. doi: 10.1038/s41467-022-35523-8. Nat Commun. 2022. PMID: 36543797 Free PMC article.
References
-
- Bizard A, Garnier F, Nadal M.. 2011. TopR2, the second reverse gyrase of Sulfolobus solfataricus, exhibits unusual properties. J Mol Biol. 4085:839–849. - PubMed
-
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M.. 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 4567224:942–945. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

