The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis-Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit
- PMID: 31504763
- PMCID: PMC6793452
- DOI: 10.1093/jxb/erz275
The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis-Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit
Abstract
Photosynthetic efficiencies in plants are restricted by the CO2-fixing enzyme Rubisco but could be enhanced by introducing a CO2-concentrating mechanism (CCM) from green algae, such as Chlamydomonas reinhardtii (hereafter Chlamydomonas). A key feature of the algal CCM is aggregation of Rubisco in the pyrenoid, a liquid-like organelle in the chloroplast. Here we have used a yeast two-hybrid system and higher plants to investigate the protein-protein interaction between Rubisco and essential pyrenoid component 1 (EPYC1), a linker protein required for Rubisco aggregation. We showed that EPYC1 interacts with the small subunit of Rubisco (SSU) from Chlamydomonas and that EPYC1 has at least five SSU interaction sites. Interaction is crucially dependent on the two surface-exposed α-helices of the Chlamydomonas SSU. EPYC1 could be localized to the chloroplast in higher plants and was not detrimental to growth when expressed stably in Arabidopsis with or without a Chlamydomonas SSU. Although EPYC1 interacted with Rubisco in planta, EPYC1 was a target for proteolytic degradation. Plants expressing EPYC1 did not show obvious evidence of Rubisco aggregation. Nevertheless, hybrid Arabidopsis Rubisco containing the Chlamydomonas SSU could phase separate into liquid droplets with purified EPYC1 in vitro, providing the first evidence of pyrenoid-like aggregation for Rubisco derived from a higher plant.
Keywords: Arabidopsis thaliana; Chlamydomonas reinhardtii; Nicotiana benthamiana; CO2-concentrating mechanism; chloroplast; photosynthesis; pyrenoid.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures
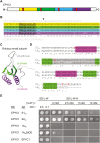


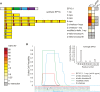

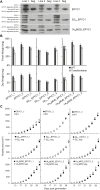

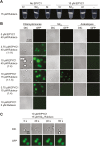
Comment in
-
How protein - protein interactions contribute to pyrenoid formation in Chlamydomonas.J Exp Bot. 2019 Oct 15;70(19):5033-5035. doi: 10.1093/jxb/erz299. J Exp Bot. 2019. PMID: 31613970 Free PMC article.
References
-
- Aigner H, Wilson RH, Bracher A, Calisse L, Bhat JY, Hartl FU, Hayer-Hartl M. 2017. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278. - PubMed
-
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. 1998. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Canadian Journal of Botany 76, 1052–1071.
-
- Bathellier C, Tcherkez G, Lorimer GH, Farquhar GD. 2018. Rubisco is not really so bad. Plant, Cell & Environment 41, 705–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

